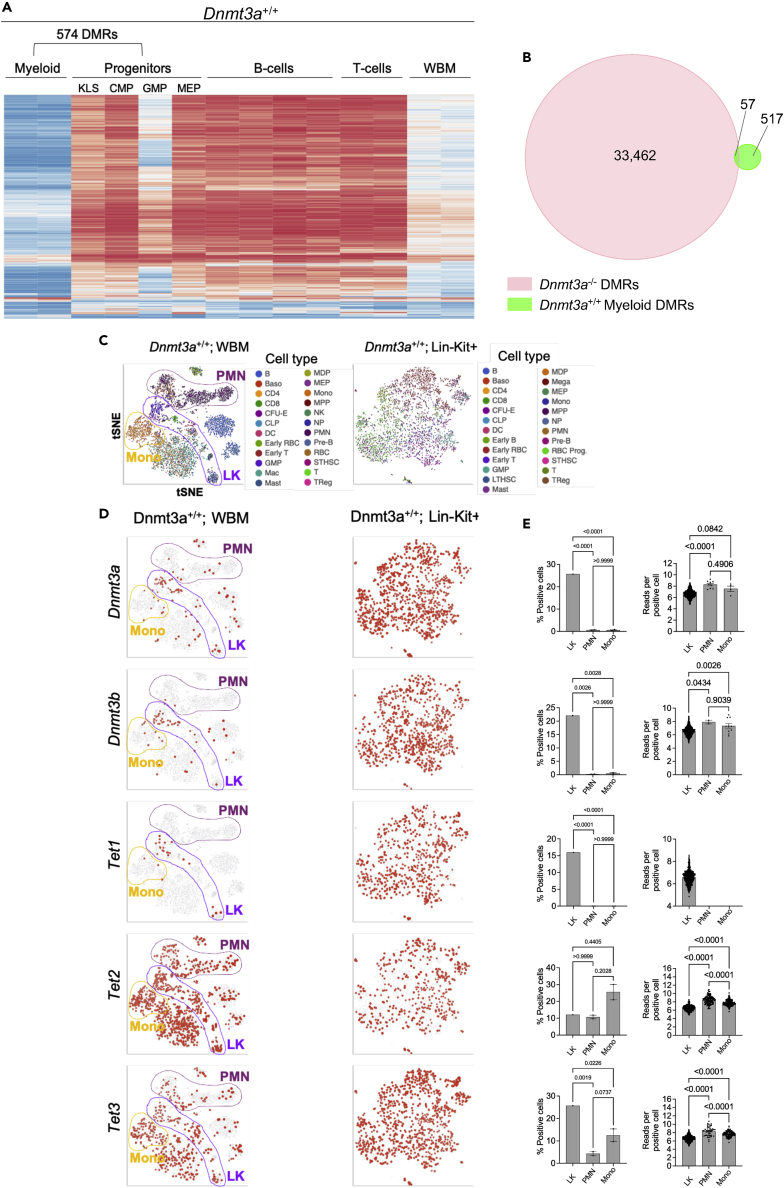
Figure 5.
Differential methylation and expression signatures in hematopoietic progenitors vs. mature myeloid cells
(A) Heatmap of mean methylation values for DMRs identified by comparing flow-purified progenitors (KLS, MEP, CMP, and GMP; grouped for analysis) and mature myeloid cells (CD11b+/Gr1+) isolated by FACS sorting. Methylation values for those DMRs in B-cells, T-cells, and whole bone marrow are passively plotted.
(B) Venn diagram showing overlap of the 33,519 DMRs defined in Dnmt3a−/− bone marrow vs. the 574 DMRs from Dnmt3a+/+ myeloid cells. Overlap between DMRs is defined as a minimum of 1 bp.
(C) tSNE projections of scRNA-seq data from whole bone marrow (left) and lineage depleted, Kit positive (Lin-Kit+) progenitor cells (right) purified by FACS. Mature neutrophils (PMN), monocytes (mono), and lineage negative, KIT positive cells (LK) are highlighted on the whole bone marrow projection.
(D) Cells are colored according to levels of expression of Dnmt3a, Dnmt3b, Tet1, Tet2, and Tet3 (top to bottom) in Dnmt3a+/+ whole bone marrow (left panels) or lineage negative c-Kit positive cells (right panels).
(E) Quantitation of percent total cells positive for expression (left panels) and number of normalized reads per positive cell (right panels) for Dnmt3a, Dnmt3b, Tet1, Tet2, and Tet3 genes in the indicated cell type. LK; lineage negative, c-KIT positive, WBM; whole bone marrow. One-way ANOVA testing was used to define statistical differences between groups. For Tet1, no reads were detected in the PMNs and Monos, so statistical comparisons could not be performed.