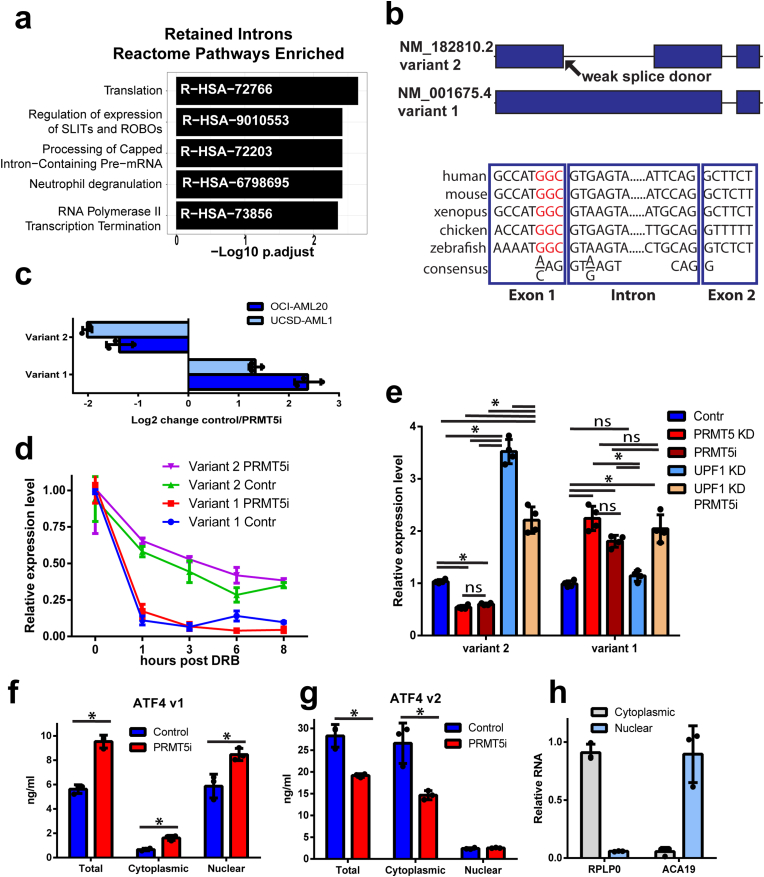
Fig. 3.
PRMT5 regulates splicing programs and ATF4 splice variant switch. a) Significantly enriched Reactome pathways in PRMT5 inhibition regulated intron retention events. b) A schematic of ATF4 gene illustrating the spliced variant 2 and retention of intron 1 that generates variant 1. The evolutionary conservation of weak 5′ splice donor sites in intron 1 of the ATF4 gene is illustrated below, red non-consensus sequence. c) Fold change in ATF4 variant 1 and 2 transcripts upon PRMT5 inhibition by 100 nM LLY283. Normalized expression fold change is shown where negative values indicate downregulation and positive – upregulation due to PRMT5 inhibition (N = 3, mean ± SD). d) Variant 1 and variant 2 transcripts have differential stability. UCSD-AML-1 cells, pretreated with LLY283 100 nM for 6 days, were exposed to 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) transcription inhibitor, and transcript levels were assessed by PCR. Normalized relative expression level is plotted, N = 3, mean ± SEM shown. e) Inhibition of nonsense-mediated decay (UPF1 knockdown) affected the variant 2 but not variant 1 transcript levels (PRMT5i, LLY283 100 nM, 6 days, UCSD-AML-1 cells). N = 3, *p < 0.05, meas±SD, two-way ANOVA. f-g) Nuclear and cytoplasmic fractions of V1 and V2 of ATF4 as determined by quantitative standard curve-based RT-PCR (N = 3, *p < 0.05, mean ± SD). h) Fractionation quality controls of nuclear ACA19 and cytoplasmic RPLP0 RNAs normalized to a nuclear or cytoplasmic fraction. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)