Figure 1.
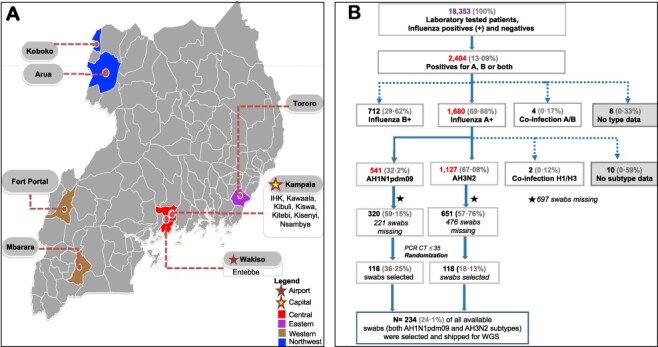
Geographical distribution of sampled sentinel sites and selection of patients’ swabs for influenza whole-genome sequencing. Panel A shows the distribution of sampled sentinel sites in the UVRI-NIC influenza surveillance programme. District hospitals included Arua-ARU, Entebbe-EBB, Fort Portal-FTL, Koboko-KBK, Mbarara-MBA, Tororo-TOR, and Kampala. Kampala had three hospitals: International Hospital Kampala-IHK, Kibuli moslem hospital-KIB, Nsambya Hospital-NSY, and four clinics: Kawaala Health Center-KIS, Kiswa Health Center-KSW, Kisenyi Health Center-KSY, and Kitebi health centre-KIS samples. Swabs were assigned unique laboratory identification numbers using the site codes and a number. For example, EBB0001 for swab 0001 from Entebbe. Panel B is a flowchart showing the exclusion and inclusion criteria for patients’ swab samples selected for influenza whole-genome sequencing (WGS). Patients diagnosed with either influenza subtypes H1N1pdm09 or H3N2 and whose swabs had a PCR CT ≤ 35 had their laboratory codes randomised based on the subtype and year of collection using the R software v3.6.3 (https://www.r-project.org For years with few swabs (n < 15), all available swabs were retrieved. The missing swabs (n = 697) consisted of some shipped to the CDC for routine surveillance and others lost due to accidental failure of a freezer. The numbers are based on the UVRI-NIC laboratory dataset only, as of 9 May 2018.
