Figure 3.
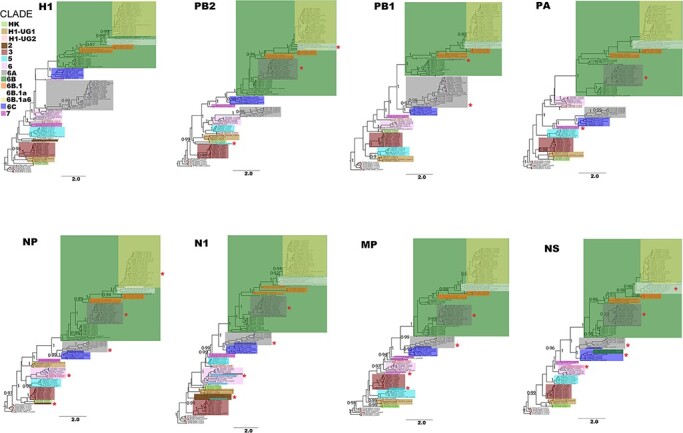
Genetic clade classification and clade switching across whole genomes of 100 H1N1pdm09 viruses sampled between 2010 and 2018 in Uganda. Viruses were classified into genetic clades based on unique amino acid substitutions in the HA1 subunit of the H1 protein(ECDC 2010). Clades in the remaining seven genes (PB2, PB1, PA, NP, N1, MP, and NS) were inferred using the H1 classified phylogeny as a reference. Original clade colours (based on the H1 phylogeny) of viruses that switched clades were maintained for visualisation purposes. Altered clades with additional virus(es) from the original clade are marked with (*). Sample nodes are coloured based on their PhyCLIP cluster ID.
