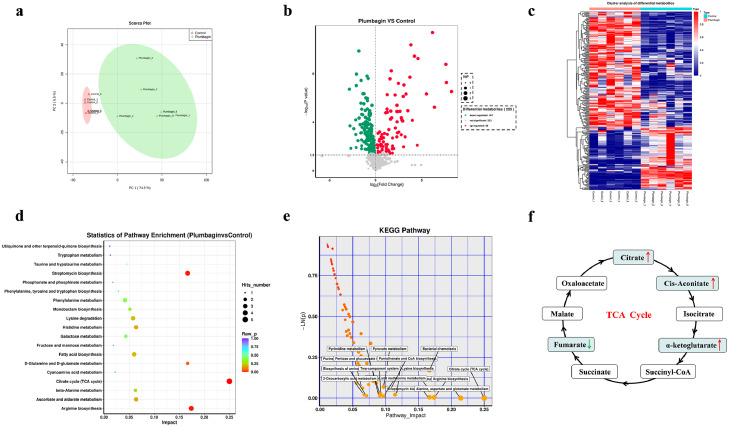
Figure 7.
Metabolomic analysis (NEG mode) of E. coli DH5α+pAM401-tet(X4) treated with plumbagin. Principal component analysis (PCA) (a) and volcano plots (b) were used to identify differential metabolites between the plumbagin-treatment group and the control group. Significantly upregulated and downregulated metabolites are shown in red and green, respectively. Non-significantly different metabolites are shown in grey. (c) Cluster analysis in NEG mode was carried out based on the heatmap, and the x- and y-axes represent the different experimental groups and the different metabolites, respectively. (d and e) KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis of metabolic pathways. The most significantly enriched pathways are shown. (f) Analysis of the abundance of metabolites in TCA cycle. The decreasing and increasing abundances of metabolites were highlighted in green and red arrows, respectively. Data are presented as the means of six biological replicates.