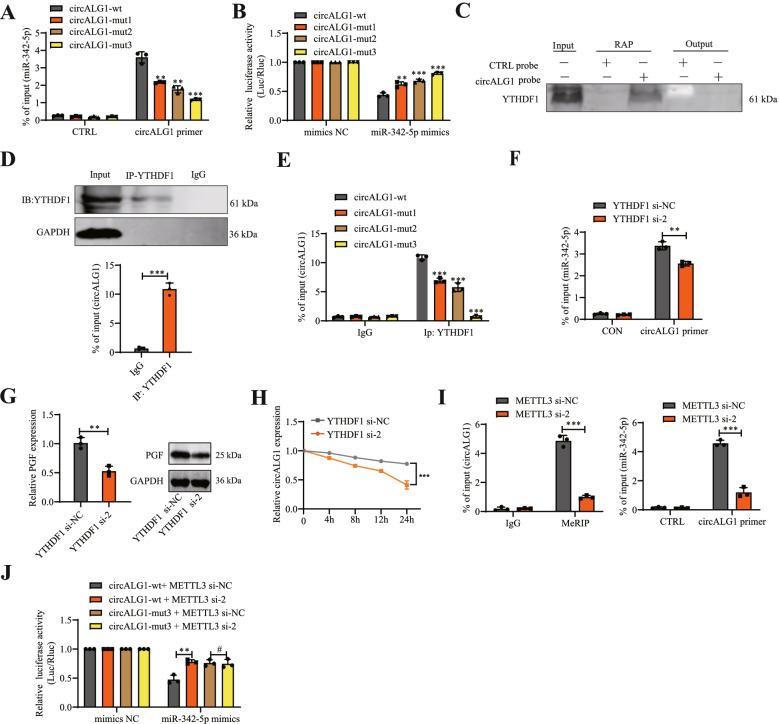
Fig. 7.
The m6A modification of circALG1 enhanced its ability to bind to miR-342-5p. A CircALG1 RAP detection of the enrichment of miR-342-5p in cells with different m6A modification levels. B The luciferase activities were measured by a dual-luciferase assay, and the Renilla/firefly luciferase light-unit ratio was calculated. C CircALG1 RAP proteins were evaluated by WB with YTHDF1 antibody. D qRT-PCR detection of the enrichment of circALG1 in the YTHDF1 immunoprecipitate obtained by RIP. E A RIP assay was used to detect the enrichment of circALG1 after mutation of the m6A modification site. F A RAP assay was used to detect the enrichment of miR-342-5p after YTHDF1 interference. G qRT-PCR and WB assays were performed to detect PGF expression levels after YTHDF1 interference. H CircALG1 stability in YTHDF1 si-NC/si-2 SW480 cells was determined by qRT-PCR after actinomycin D treatment for the time indicated. I MeRIP analysis of the circALG1 m6A modification levels (left) and miR-342-5p enrichment after METTL3 interference, as detected by circALG1 RAP (right). J The luciferase activities were measured by a dual-luciferase assay, and the Renilla/firefly luciferase light-unit ratio was calculated. The results are presented as the mean ± s.d. and are representative of at least 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, #p > 0.05