Figure 3. FBXO7 protects EYA2 from SCFFBXW7-dependent degradation.
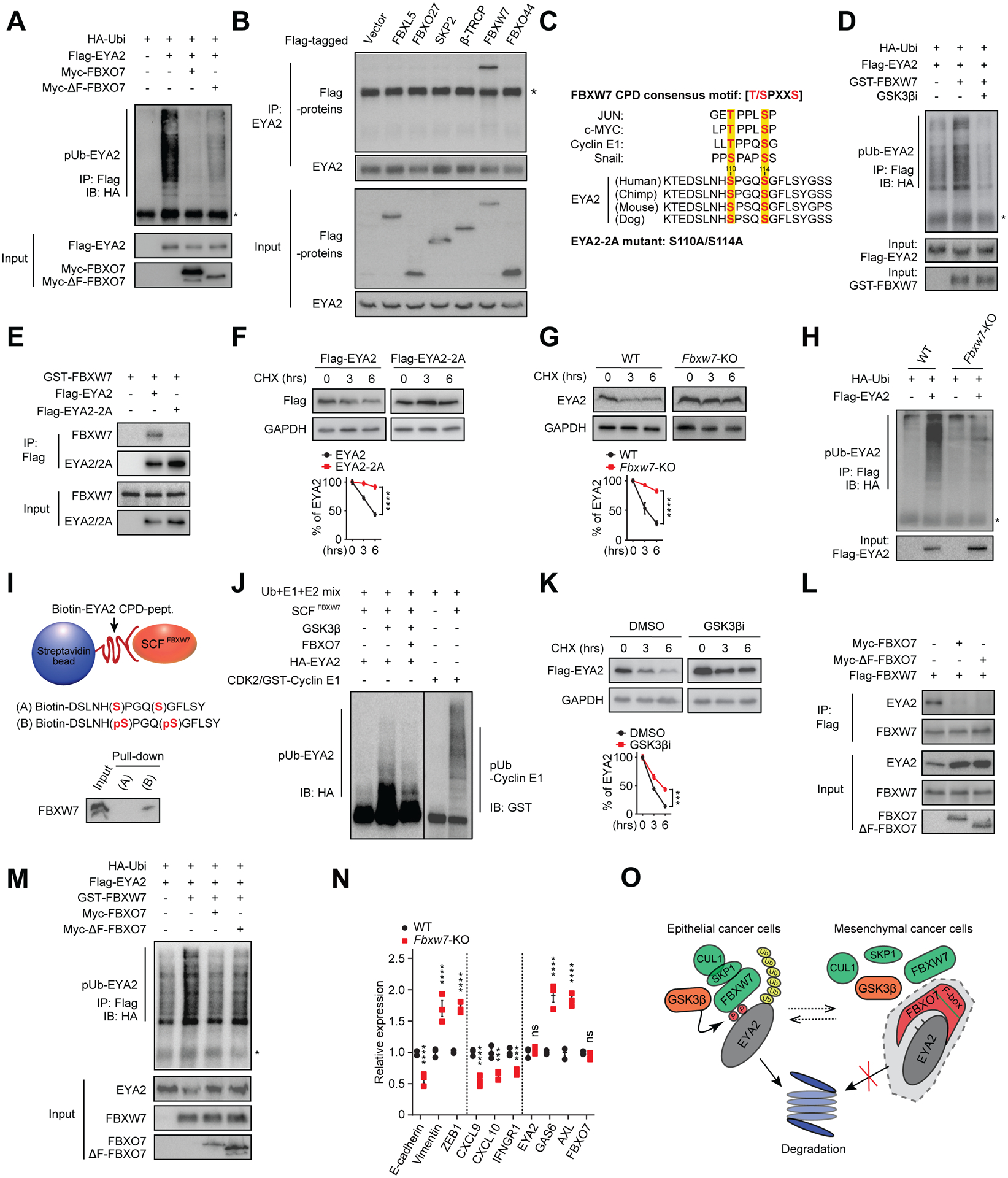
(A) Immunoblot of polyubiquitylated (pUb) Flag-EYA2 in HEK293T cells transfected with the indicated vectors. Input proteins are shown. *, IgG heavy chain.
(B) Co-IP of endogenous EYA2 with the indicated F-box proteins. *, IgG heavy chain.
(C) Putative EYA2 CPD motif across species and compared with other SCFFBXW7 substrates. GSK3β phosphorylated residues are highlighted.
(D) Immunoblot of pUb Flag-EYA2 in HEK293T cells transfected with the indicated vectors and treated with GSK3βi. Input proteins are shown. *, IgG heavy chain.
(E) Co-IP of WT and CPD mutant Flag-EYA2–2A with GST-FBXW7.
(F and G) WT and Flag-EYA2–2A stability in MDA-MB-231 cells (F) and endogenous EYA2 in WT and Fbxw7 KO HCT116 cells (G) (top panels). GAPDH, control. Relative signal intensities (bottom panels) (n = 3).
(H) Immunoblot of pUb Flag-EYA2 in WT and Fbxw7 KO HCT116 cells. Input shown.
(I) In vitro binding of SCFFBXW7 with un-phosphorylated (A) or phosphorylated (B) EYA2 CPD peptide.
(J) In vitro ubiquitylation reactions of SCFFBXW7 with EYA2 with/without FBXO7. Cyclin E1, positive control.
(K) Flag-EYA2 stability in MDA-MB-231 cells treated with vehicle (DMSO) or GSK3βi (top panel). GAPDH, control. Relative signal intensities (bottom panel) (n = 3).
(L) Co-IP of Flag-FBXW7 with endogenous EYA2 in MDA-MB-231 cells transfected with the indicated FBXO7 vectors.
(M) Immunoblot of pUb Flag-EYA2 in HEK293T cells transfected with the indicated vectors. Input proteins shown.
(N) RT-qPCR analysis of indicated genes in WT and Fbxw7 KO HCT116 cells (n = 3).
(O) Model of FBXO7 inhibition of GSK3β-SCFFBXW7-mediated degradation of EYA2.
Data represent mean ± SEM. ns, not significant; ***p < 0.001; ****p < 0.0001 by two-way ANOVA followed by Sidak’s multiple comparisons test (F), and Sidak’s multiple comparisons test (G, K, and N).
See also Figure S3.
