Fig. 5. MCRS1 regulates histone acetylation of BA transporter genes.
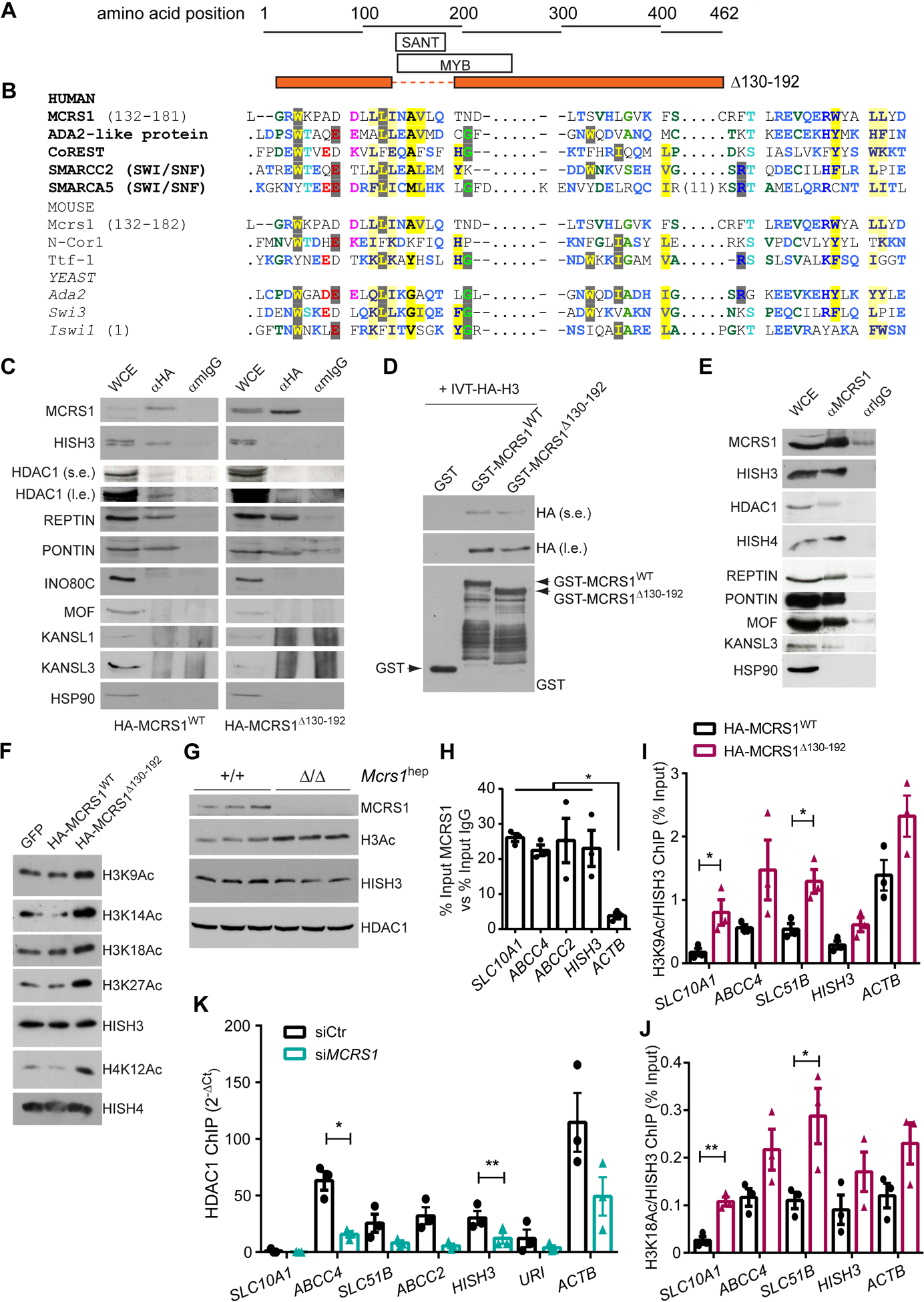
(A) Schematic representation of the protein sequence of MCRS1Δ139–192 mutant. (B) Sequence alignment of human, mouse and yeast MCRS1 protein sequence with proteins containing a SANT domain. (C) IP performed in HepG2 cells overexpressing HA-MCRS1WT and HA-MCRS1Δ130–192 mutant proteins (N=3). s.e. and l.e. mean short exposure and long exposure, respectively. (D) WB performed in preparations of in vitro translated HA-HISH3 incubated with GST-, GST-MCRS1WT and GST-MCRS1Δ130–192. (E) IP performed in HepG2 cells (N=3). (F) WB of indicated proteins of core histones isolated from HepG2 cells overexpressing GFP, HA-MCRS1WT and HA-MCRS1Δ130–192 mutant proteins (N=3). (G) WB performed in isolated hepatocytes from 6-week-old Mcrs1(+/+)hep and Mcrs1(Δ/Δ)hep mice. (H) Chromatin IP (ChIP) using MCRS1 antibody performed in HepG2 cells (N=3). (I) and (J) ChIPs using H3K9Ac (I) and H3K18Ac (J) antibodies performed in HepG2 cells overexpressing HA-MCRS1WT and HA-MCRS1Δ130–192 mutant proteins (N=3). (K) ChIP using HDAC1 antibody performed in HepG2 cells transfected with siCtr or siMCRS1 (N=3). Statistical analysis was performed using two-way ANOVA with Tukey’s multiple comparisons test in (H), and unpaired two-tailed Student’s t test in (I) to (K). Data are represented as means±SEM. *, p≤0.05; **, p≤0.01.
