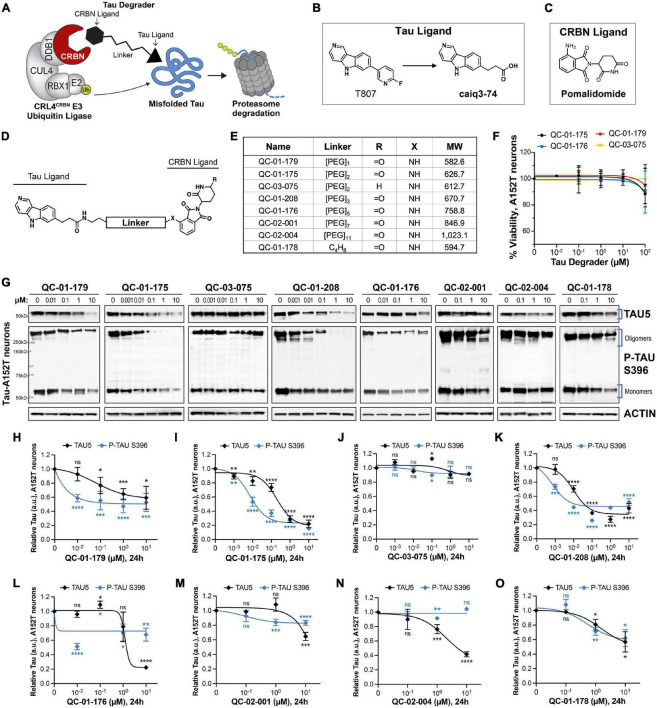
FIGURE 1.
Design strategy for CRBN-recruiting degraders and concentration effect on tau and P-tau of human FTD neurons. (A) Working model for hetero-bifunctional tau degraders designed to preferentially recognize pathological forms of tau (misfolded protein) and simultaneously engage with CRBN (CRL4CRBN E3 ubiquitin ligase complex). Formation of the ternary complex is expected to enhance tau ubiquitination and degradation by the proteasome. (B–D) Degraders were synthesized based on the T807 core scaffold for tau recognition (B), a thalidomide analog as an E3 ligand (pomalidomide, C) for CRBN engagement, and a variable linker length and composition to maximize target clearance efficiency (D). (E) Summary of the chemical properties of CRBN-recruiting degrader molecules of the QC-Series. (F) Neuronal viability of tau-A152T neurons at 6 weeks of differentiation treated with representative degraders of the series for 24 h. Viability is shown relative to vehicle-treated neurons (100%) and each data point represents the mean% viability ± SD, n = 3. (G–O) Concentration effect of degraders on tau protein of A152T neurons (6-week differentiated) by analysis of total tau (TAU5) and P-tau S396 levels upon treatment for 24 h. Representative western blots are shown (G) and brackets indicate bands quantified for TAU5 and P-tau S396 densitometry (H–O). Data points represent mean densitometry normalized to actin and relative to vehicle ± SEM (n ≥ 3). Data points for QC-01-175 (I) and QC-03-075 (J) include values from new biological replicates averaged with analysis previously published (Silva et al., 2019). The 0 μM degrader data points correspond to vehicle alone (DMSO) control treatment. Statistics: 2-tailed unpaired Student’s t-test for each concentration relative to vehicle; nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.