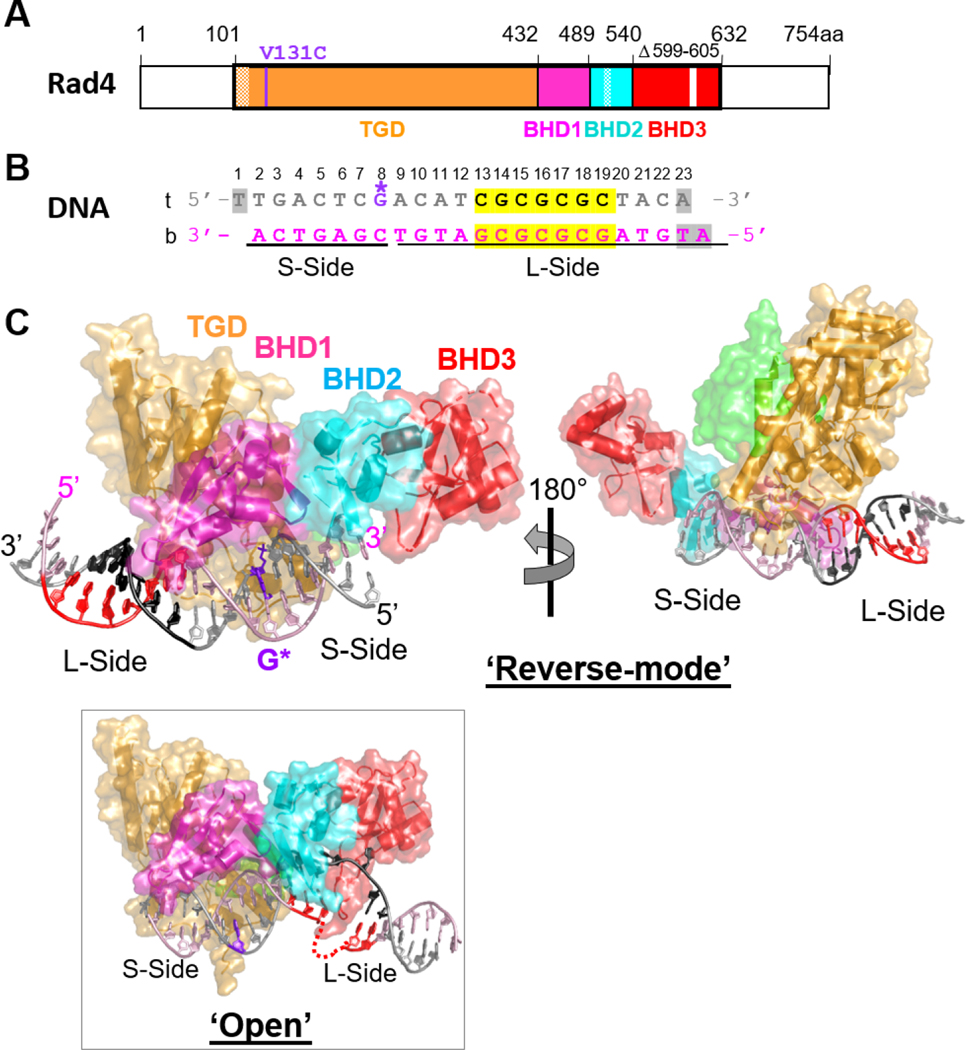
Fig.1. Crystal structure of the Δβ-hairpin3 mutant Rad4–Rad23 complex tethered to DNA containing alternating CG/GC repeat shows ‘reverse mode’ binding.
(A) The crystallized Rad4 construct spans residues 101–632. The transglutaminase domain (TGD) is indicated in orange, β-hairpin domain 1 (BHD1) magenta, BHD2 cyan and BHD3 red. The deleted region in the BHD3 β-hairpin (residues 599–605) is indicated in white. The disordered regions in crystals (residues 101–128, 518–525) are checkered. The V131C point mutation introduced for disulfide crosslinking is in purple. (B) The 23-bp CGC/GCG DNA construct for crystallization. Top strand (‘t’) is in silver and the bottom (‘b’) in pink. The CG/CG repeats are highlighted in yellow and colored black in ‘t’ and red in ‘b’. The disulfide-modified nucleotide, G* in dG8 is shown in purple. The DNA residues with missing electron densities are shaded in gray. The bottom strand was the damage-containing strand in the ‘open’ structures of lesion-bound Rad4 (PDB ID: 2QSG, 6CFI). (C) The ‘reverse- mode’ structure of the Δβ-hairpin3 mutant bound to CGC/GCG DNA duplex (PDB ID: 6UG1). The color scheme is the same as in (A) and (B). Rad23’s Rad4-binding domain (R4BD) is shown in light green. The right panel shows the structure rotated by 180 ° along a vertical axis. (Inset) The ‘open’ structure previously determined with CCC/GGG DNA tethered to the WT Rad4 complex (PDB ID: 4YIR).