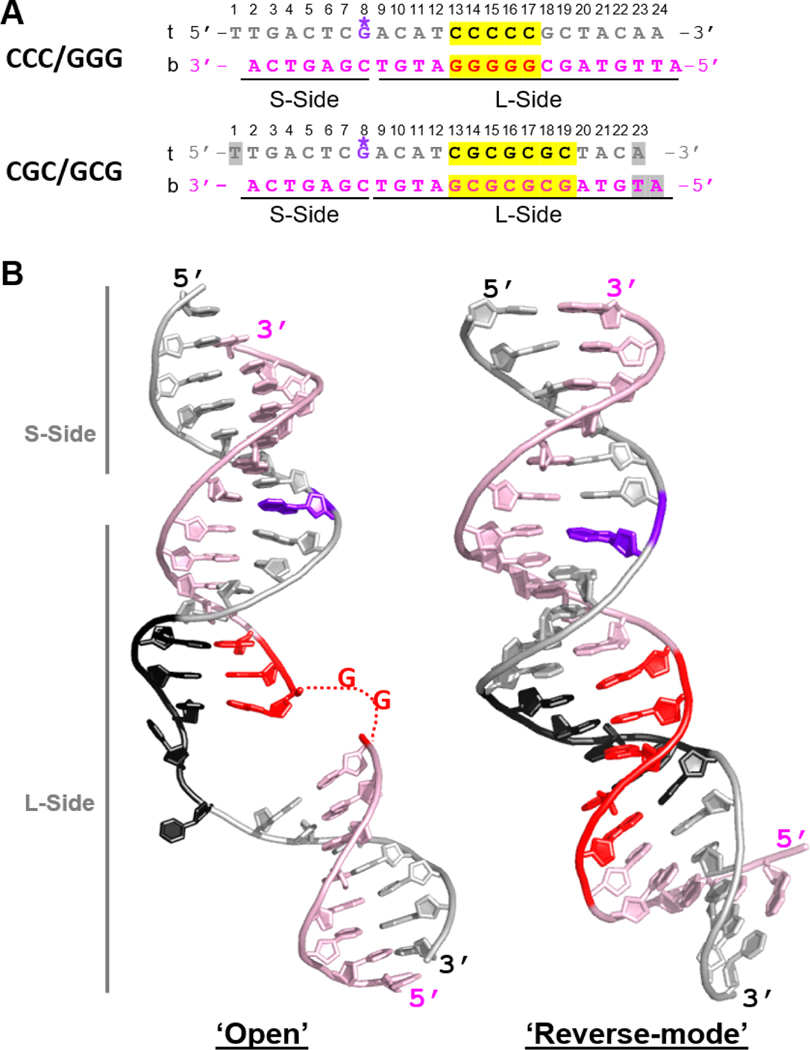
Fig.2. Comparison of DNA conformations in ‘open’ versus ‘reverse mode’ structures.
(A) The sequences of the CCC/GGG DNA that forms ‘open’ or ‘open-like’ structures when tethered to Rad4 and of the CGC/GCG DNA forming a ‘reverse mode’ structure. The top (‘t’) and bottom strands (‘b’) are in silver and light pink, respectively; consecutive C/G’s and alternating CG/GC repeats are highlighted in yellow and colored black in ‘t’ and red in ‘b’. The DNA residues with missing electron densities are shaded in gray. The crosslinkable G* is in purple. The short (S-side) and long sides (L-side) of the DNA duplexes are designated with respect to the G* tethering site. (B) (left) The ‘open’ DNA conformation in the WT Rad4CCC/GGG structure (PDB ID: 4YIR); (right) The ‘reverse mode’ DNA shown in the Δβ-hairpin3-CGC/GCG DNA structure (PDB ID: 6UG1).