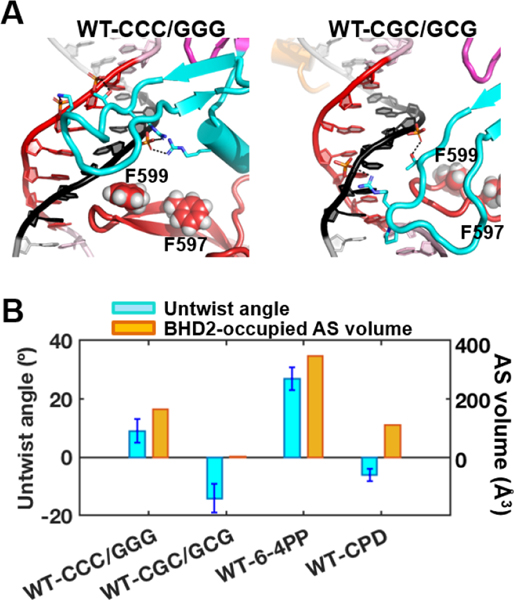
Fig.6. Initial binding of Rad4 to the CCC/GGG and CGC/GCG sequence-containing duplexes.
(A) Best representative structures upon initial binding of Rad4. The structures are shown in cartoon representation color-coded as in Figure 1. Heavy atoms of the BHD2 amino acid side chains and the potential open site’s DNA backbone phosphate groups that form hydrogen bonds are shown in sticks. Hydrogen bonds are shown as black dashed lines. Side chains of two Phe (F599 and F597) in the β−hairpin3 are shown in spheres. The structure for the CCC/GGG case is adapted from Figure 1A in [28]. (B) Potential ‘open’ site untwisting (6-mer) and BHD2 binding into the minor groove were quantified using the untwist angles and the BHD2-occupied AS volumes in the minor groove for the Rad4 binding. The values for the CCC/GGG sequence are from [28], and for the 6–4PP and CPD, they are from [22]. The CCC/GGG sequence shows significant BHD2 binding and modest untwisting, resembling the well-recognized and repaired 6–4PP. In contrast, the CGC/GCG sequence shows no BHD2 binding and slight over-twisting, reminiscent of the poorly recognized/repaired CPD. The standard deviations of block averaged means [110, 111] for the untwist angles are shown. Full details of the block averaging method are given in SI Methods.