Figure 2.
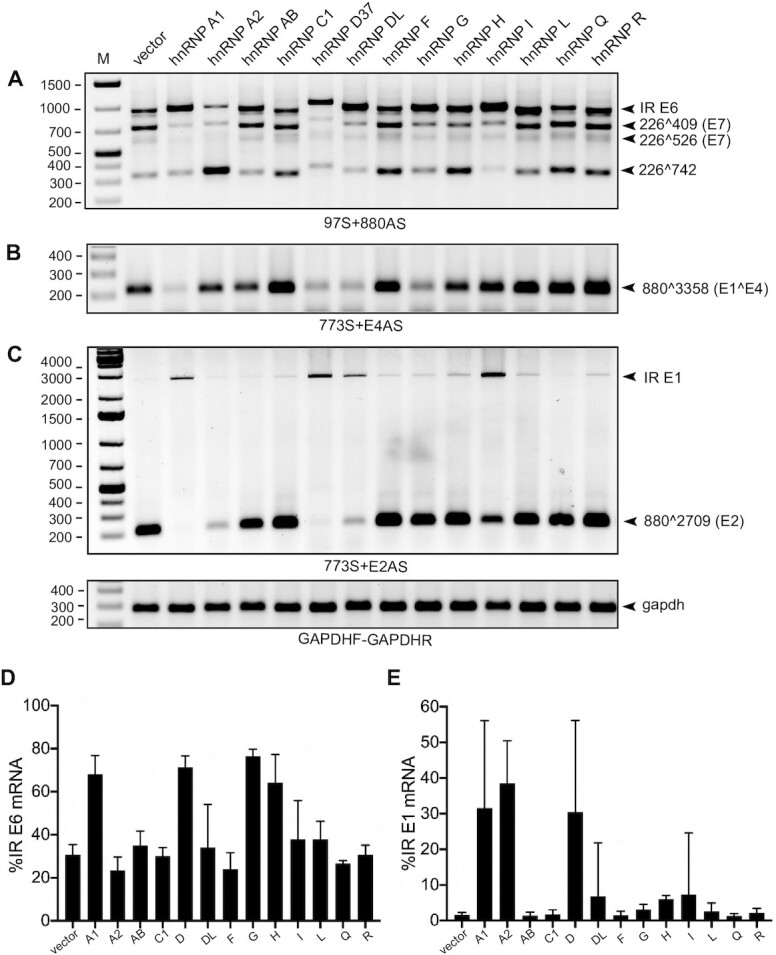
Identification of hnRNP D as a strong suppressor of HPV16 E6E7 and E1E2 mRNA splicing. (A–C) The effect of 13 different hnRNPs on HPV16 alternative mRNA splicing was determined by RT-PCR with different pairs of primers. HPV16 subgenomic plasmid pC97ELsLuc (Figure 1) was cotransfected individually with each of 13 different hnRNP expressing plasmid into HeLa cells. p37 isoform of hnRNP D (hnRNP D37) was used. Total RNA was extracted at 24 hpt and the various spliced HPV16 mRNAs were monitored by RT-PCR using HPV16-specific RT-PCR primer pairs: (A) E6/E7 mRNAs: 97S+880AS, (B) E4 mRNA: 773S+E4AS and (C) E1/E2 mRNAs: 773S+E2AS. Resulting RT-PCR products were electrophoresed on agarose gels and gel images are displayed. The schematic structures of the HPV16 spliced mRNAs and the locations of the HPV16-specific RT-PCR primers are indicated in Figure 1. Primer sequences are available in Supplementary Table S1. Each band was verified by sequencing and HPV16 spliced mRNA isoforms are indicated to the right of each gel. Validation of RT-PCR was also evaluated by reverse transcriptase negative samples using corresponding RNA samples. M: DNA size marker. Representative gel images of results repeated at least three-times are shown. (D) The effect of each hnRNP protein on the enhancement of HPV16 intron-retained (IR) E6 mRNA production shown in (A) was evaluated. A band intensity of each spliced isoform in (A) was quantitated described in Materials and Methods. The percentage of intron-retained E6 mRNA over a total sum of all four isoforms (intron-retained (IR) E6-, 226∧409-, 226∧526-, and 226∧742-mRNA) was calculated. (E) The effect of each hnRNP protein on the enhancement of HPV16 intron-retained E1 mRNA production shown in (C) was evaluated as described in (D).
