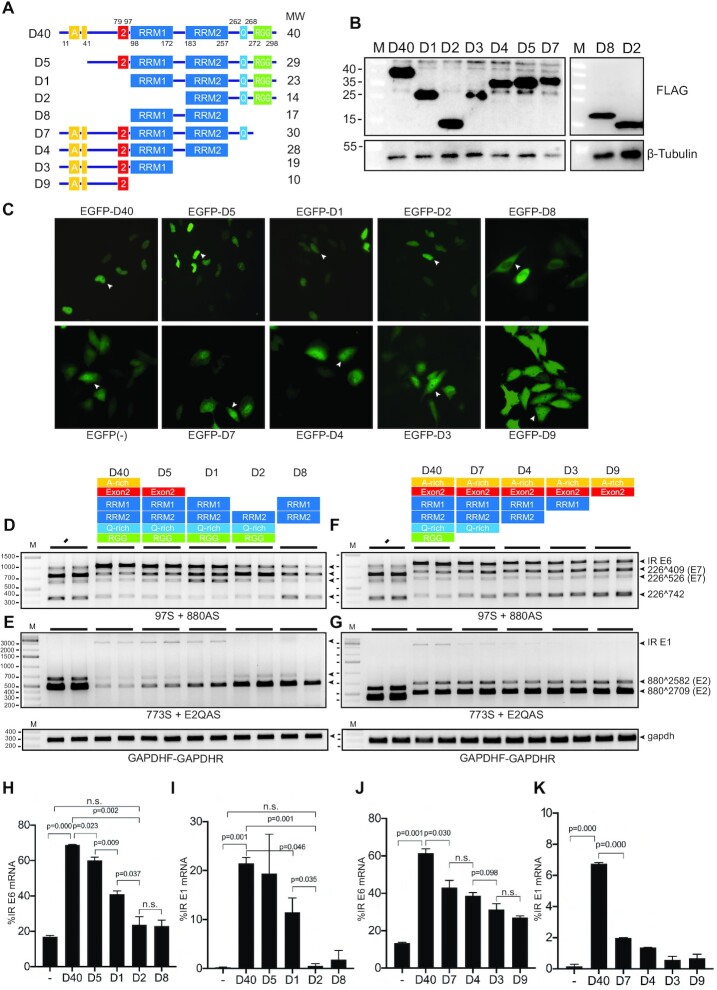
Figure 4.
RRM1 domain of hnRNP D40 was required for HPV16 E6 and E1 intron retention whereas N-terminal and C-terminal domains contributed to modification of HPV16 early alternative mRNA splicing. (A) Schematic structures of wild-type hnRNP D40 and hnRNP D40 deletion mutants. All mutants were fused with FLAG-tag at its N-terminus. Box with A: Alanine(A)-rich region, box with 2: exon 2 region, box with RRM: RNA recognition motif (RRM) domain, box with Q: Glutamine(Q)-rich region and box with RGG: Arginine-Glycine/Arginine-Glycine-Glycine (RG/RGG) motif region. The names of each deletion mutant are indicated to the left and the predicted molecular weight of each protein is indicated to the right. (B) Western blotting against the FLAG epitope demonstrating protein expression of the hnRNP D40 deletion mutants depicted in (A). M: molecular weight marker. (C) Subcellular localization of indicated deletion mutants of hnRNP D40. Each mutant was fused to the C-terminus of EGFP. Representative cells expressing each mutant are highlighted by white arrowheads and shown in Supplementary Figure S3 with higher magnification. (D–G) Effect of FLAG-hnRNP D40 deletion mutants on HPV16 mRNA splicing was monitored by HPV16 RT-PCR on RNA extracted from HeLa cells transfected in the absence (−) or presence of hnRNP D40 or deletion mutants thereof. Representative gel images from experiments independently repeated three times are shown. (H and J) Percentage of intron-retained E6 mRNA over all four E6/E7 alternatively spliced isoforms quantified from (D) or (F). (I and K) Percentage of intron-retained E1 mRNA percentage over all three E1/E2 alternatively spliced isoforms quantified from (E) or (G). Proportions among all four isoforms is shown in Supplementary Figures S3B and S3C. Student’s t-test was executed and obtained P values were displayed; n.s., no significance.