Figure 9.
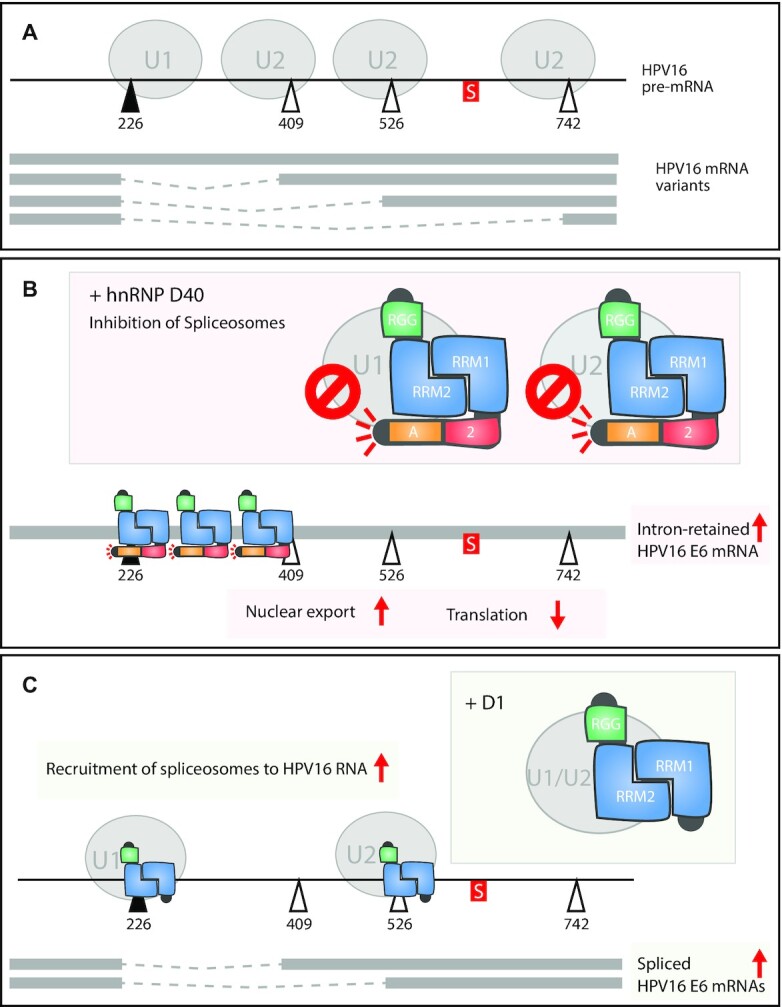
Model how hnRNP D regulates HPV16 E6/E7 mRNA splicing. (A) In regular condition, four splice isoforms of HPV16 E6/E7 mRNAs are produced. This is regulated by fine-tuned recruitment of cellular U1 spliceosome factor to HPV16 E6/E7 splice donor SD226 and that of U2 spliceosome factors to HPV16 E6/E7 splice acceptors SA409, SA526 or SA742. (B) hnRNP D40 binds to both U1 spliceosome factor and U2 spliceosome factor at its C-terminus and with some extent at its N-terminus, which inhibits spliceosome factor activity. This inhibitory function is mediated by N-terminus of hnRNP D. At the same time, hnRNP D interacts with several sites in HPV16 E6/E7 coding RNAs at its RRM1/RRM2 domain. As a result, hnRNP D mainly inhibits 226∧409 splicing on the HPV16 E6/E7 mRNAs. The intron-retained E6 mRNAs are accompanied by hnRNP D to the cytoplasm. However, hnRNP D association on intron-retained E6 mRNA suppresses translation of E6 protein by unknown mechanism. To enhance translation of HPV16 E6 protein from intron-retained E6 mRNA, hnRNP D40 must need to be replaced from intron-retained E6 mRNA. (C) Deletion mutant of hnRNP D40, D1 that doesn’t possess N-terminal A-rich and exon 2 but harbors intact RRM1/RRM2 domain and C-terminus, still has potential to interact with spliceosome factors and with HPV16 RNA though it loses inhibitory effect on spliceosome factors. As a result, D1 mutant mediates recruitment of spliceosome factors to HPV16 splice sites, thereby increasing HPV16 E6/E7 mRNA splicing.
