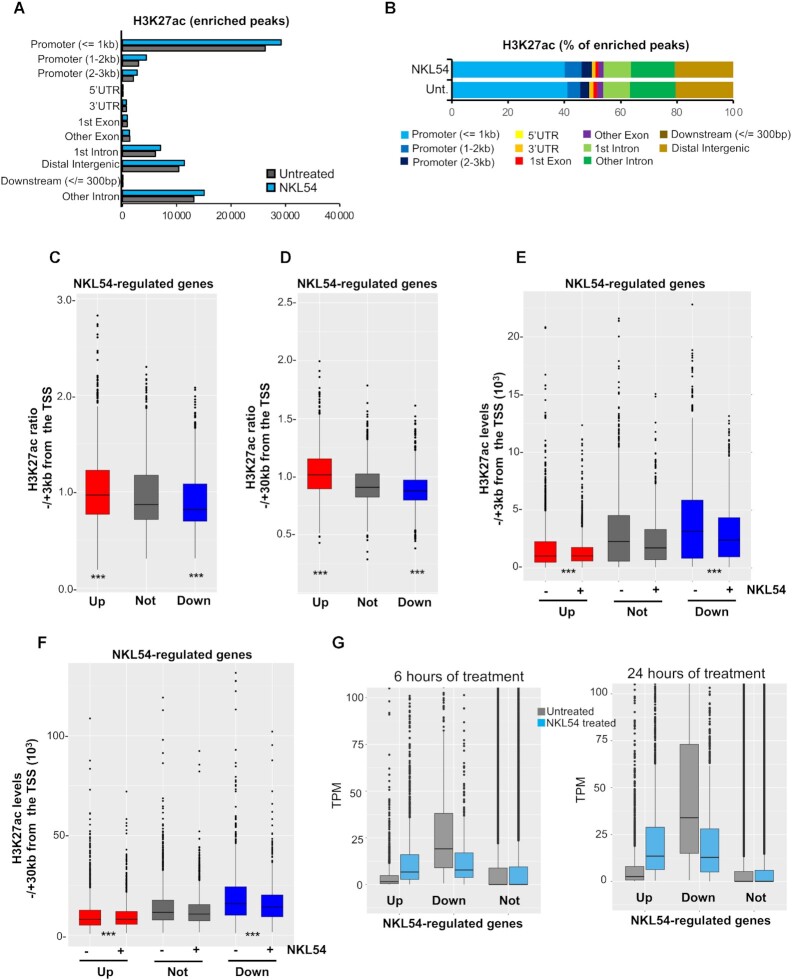
Figure 7.
Variation of H3K27ac distribution at the genomic level. (A) Genomic distribution of the H3K27ac-enriched IDR-defined peaks identified by MACS2 in SK-UT-1 cells treated (n = 72 765) or untreated (n = 64 232) for 14 h with 5 μM NKL54. (B) As in panel A, with values represented as percentages. (C) H3K27ac ratio between NKL54 treated and untreated cells within ±3 kb from TSS. ChIP-seq data are from experiment 1. Genes not-regulated by NKL54 were selected based on having the lowest combined gene expression variations at 6 and 24 h from treatment. The boxes indicate the interquartile range with the center line representing the median value. The outliers are plotted as dots. (D) As in panel C, with H3K27ac ratio between NKL54 treated and untreated cells calculated within ±30 kb from TSS. (E) Overall acetylation level in the ± 3kb region centered on gene TSS of the indicated gene categories in presence or absence of NKL54. Boxes plotted as in panel C. (F) As in panel E with overall acetylation level measured within a ±30 kb region centered on gene TSS of the indicated gene categories in presence or not of NKL54. (G) TPM values are shown after treatment or not with NKL54 for the respective times. TPM measure was calculated from a gene model where isoforms were collapsed into a single gene. Significances were tested using the Mann–Whitney U test.