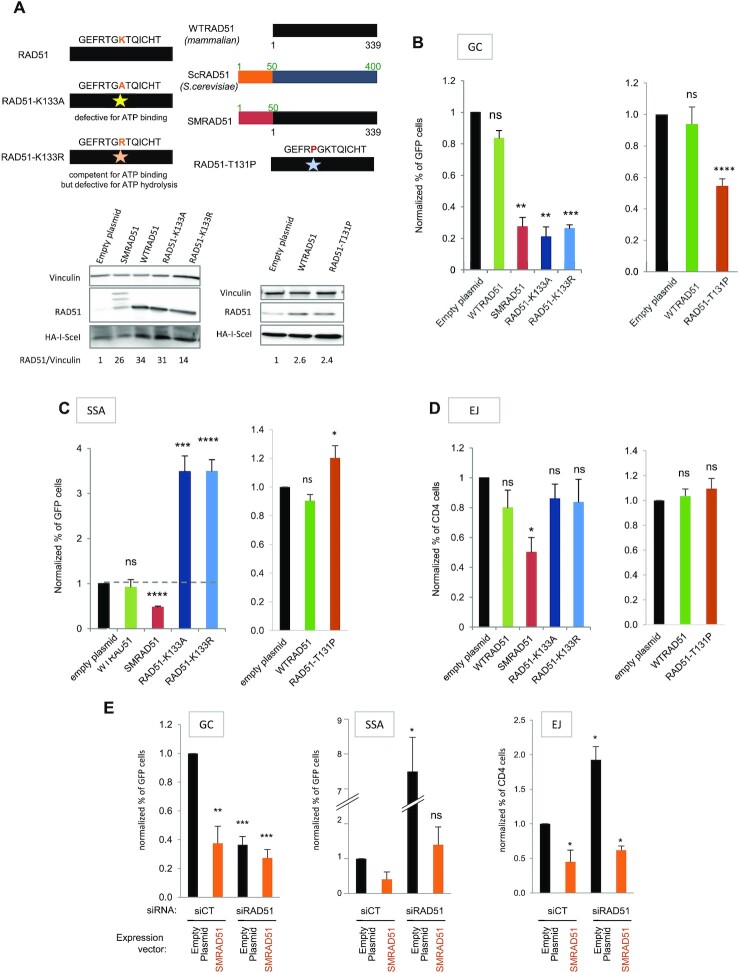
Figure 3.
Impact of the different RAD51 dominant negative forms on DSB repair. (A) The different RAD51 dominant negative forms: ATPm-RAD51s mutated on the K133 ATP binding site, SMRAD51 chimera; WTRAD51 (mammalian), Saccharomyces cerevisiae ScRAD51 and the chimera SMRAD51 are aligned, revealing a block of 50 N-ter amino acids (AAs) present in scRAD51 but absent from WTRAD51. SMRAD51 corresponds to the fusion of 50 N-ter aminoacids from yeast to full-length WTRAD51 (11). RAD51-T131P corresponds to a mutation described in a patient suffering from Fanconi anemia group R (49). Lower panel: Expression of the different RAD51 dominant-negative forms. (B–D) Impact of the different RAD51 dominant-negative forms on GC (B), SSA (C) and EJ (D). The values are shown normalized to the control transfected with an empty vector (in black) and represent the average ± SEM of at least 5 independent experiments. (E) Impact of mixed endogenous RAD51/SMRAD51 on gene conversion, SSA and EJ. SMRAD51 was expressed in cells with silenced endogenous RAD51. The values are shown normalized to the control siRNA (in black) and represent the average ± SEM of at least 5 independent experiments. (*P< 0.05, **P< 0.01, ***P < 0.001, ****P< 0.0001, t-test compared to ‘empty plasmid’).