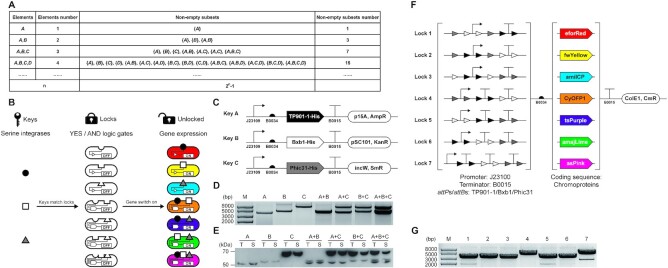
Figure 1.
Design and validation of the SYMBIOSIS framework. (A) Schematic of the mathematical ‘set’ concept. Nonempty subsets are organized with elements. The number of nonempty subsets increases exponentially with the element number (n) as 2n − 1. (B) Design of the KmL model. Three orthogonal serine integrases (TP901-1: black circle; Bxb1: white square; and Phic31: gray triangle) work as ‘keys’ to match E. coli strains (ovals with gaps) containing seven different ‘lock’ genetic circuits. A correct ‘key–lock’ match can unlock the closed circuit and switch on chromoprotein expression. (C) Architectures of three ‘key’ plasmids. TP901-1, Bxb1 and Phic31 are constructed in three compatible plasmids, respectively, with a weak constitutive promoter J23109 (pFB1–pFB3, see Supplementary Table S4). (D) Compatibility of single, double or triple ‘key’ plasmids in one E. coli strain. Extracted plasmids are digested by XbaI and then separated by agarose gel electrophoresis. (E) Western blot analysis of individual expression and co-expression of TP901-1 (56.5 kDa), Bxb1 (57.2 kDa) and Phic31 (67.9 kDa) labeled with the anti-His antibody. T, total protein; S, soluble protein. (F) Schematic of seven ‘lock’ plasmids. ‘Lock’ biobricks (left brackets) are assembled with a strong promoter J23100, a strong terminator B0015 and attP–attB sites (TP901-1 attP/attB: black triangles; Bxb1 attP/attB: white triangles; and Phic31 attP/attB: gray triangles). Seven different chromoproteins (eforRed, fwYellow, amilCP, CyOFP1, tsPurple, amajLime and asPink, right brackets) work as output signals. The seven ‘lock’ plasmids are constructed using the same vector that is compatible to three ‘key’ plasmids. (G) Plasmid sizes of the seven ‘locks’ (pFB20–pFB26, see Supplementary Table S5) after XbaI digestion. Expected bands of the seven digested plasmids are observed at 3371, 3401, 3356, 3521, 3506, 3509 and 3647 bp, respectively.