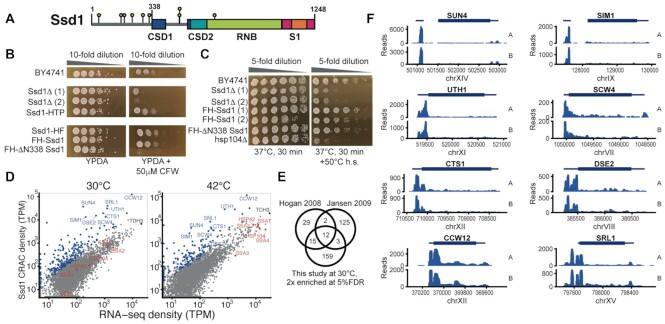
Figure 1.
Ssd1 binds to 5′ untranslated regions (5′UTRs) of its target transcripts in vivo. (A) Domain overview of Ssd1. Boxes indicate folded domains with separating grey lines indicating natively unstructured regions; yellow lollipops indicate phosphorylation sites of Ssd1. (B) Wild-type and Ssd1 mutant yeast strains grown at 30°C on YPDA [yeast extract/peptone/dextrose (glucose)/adenine] without or with 50 μM calcofluor white (CFW). (C) Wild-type and Ssd1 or Hsp104 mutant strains grown overnight at 30°C and then incubated either at 37°C for 30 min or using an induced thermotolerance protocol. (D) Ssd1-bound CRAC (cross-linking and analysis of cDNAs) read density compared to RNA-seq reads in transcripts per million (TPM, mean over two biological replicates), aligned to full-length transcripts including annotated UTRs. Selected Ssd1 targets are highlighted in blue and selected heat-induced transcripts in red. (E) Comparison of Ssd1-bound mRNAs reported by CRAC analysis with previous RNA immunoprecipitation and microarray studies that were also conducted in rich media at 30°C. We conservatively report transcripts that are 4-fold enriched in Ssd1 CRAC reads compared to RNA-seq, and with at least 20 TPM in the RNA-seq data. (F) Unnormalized CRAC read counts (pileups) on selected Ssd1-bound transcripts from two biological replicates at 30°C, aligned to the yeast genome, with 5′UTRs oriented on the left.