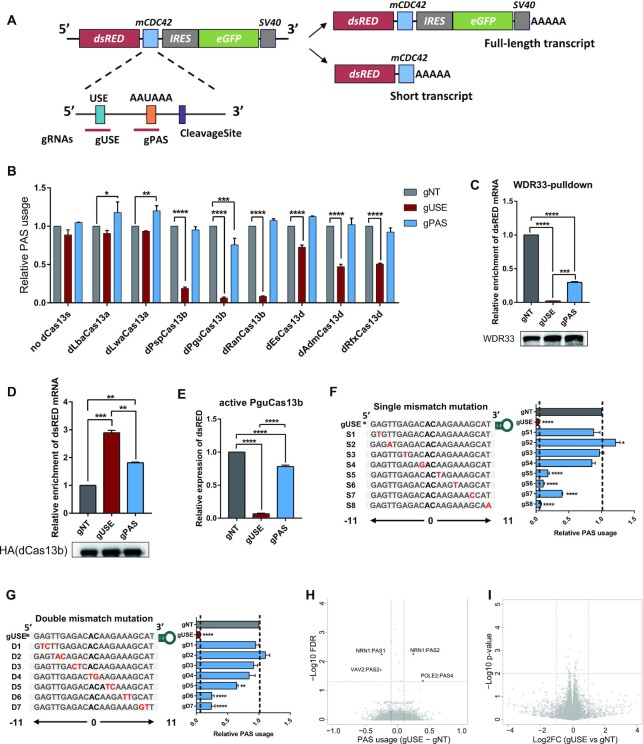
Figure 1.
Repression of PAS usage with different dCas13 proteins. (A) Illustration of the reporter gene used to examine APA regulation. A 232-nucleotide (nt) sequence flanking an endogenous PAS from mouse gene cdc42 was inserted. dsRED, red fluorescent protein; IRES, internal ribosome entry site; EGFP, enhance green fluorescent protein; AAAAA, poly(A) tail. (B) Relative usage of the target PAS in HEK293TAPA reporter cells transfected by one non-targeted control gRNA (gNT) and two gRNAs targeting at the PAS signal (gPAS) or USE (gUSE), respectively, together with or without different dCas13 proteins. Relative PAS usage was calculated by the abundance ratio between the two regions (dsRED/EGFP) measured using RT-qPCR and normalized to non-targeted gRNA (gNT). Without dCas13 proteins, both gRNAs showed no effect on the PAS usage, while six out of eight dCas13 proteins, dPspCas13b, dPguCas13b, dRanCas13b, dEsCas13d, dAdmCas13d and dRfxCas13d significantly reduced the PAS usage, when co-transfected with the USE-targeting gRNA (gUSE). Among the six effective dCas13 proteins, dPguCas13b showed the highest efficiency. Compared to gUSE, gRNA targeting PAS signal (gPAS) showed a much milder inhibitory effect. (C) RIP assays against WDR33 were performed in HEK293TAPA reporter cells with dPguCas13b and indicated gRNAs co-transfected, and the enrichment of dsRED mRNA was measured using RT-qPCR and normalized to the input total mRNA. The targeted reporter mRNA bound by WDR33 was significantly decreased in cells transfected with dPguCas13b and gUSE, compared to that in cells transfected with non-targeting control gRNA (gNT). Compared with gUSE, the decrease of WDR33 bound reporter mRNA was much subtler in cells transfected with gPAS. The immune-precipitated WDR33 protein in each RIP experiment was measured by Western Blot, as indicated in the lower panel. (D) RIP assays against HA-tagged dPguCas13b were performed in HEK293TAPA reporter cells co-transfected with HA-tagged dPguCas13b and indicated gRNAs, and the enrichment of dsRED mRNA was measured using RT-qPCR and normalized to the input mRNA. The targeted reporter mRNA bound by dPguCas13b was significantly higher in cells transfected with dPguCas13b and gUSE, compared to that in cells transfected with non-targeting control gRNA (gNT). Compared with gUSE, the enrichment of dPguCas13b bound reporter mRNA was significantly lower in cells transfected with gPAS. The protein level of dPguCas13b used in each RIP experiment was measured by Western Blot, as indicated in the lower panel. (E) Abundance of the reporter gene mRNA in HEK293TAPA reporter cells transfected with active PguCas13b and indicated gRNAs. The level of dsRED mRNA was measured using RT-qPCR and normalized to that of GAPDH. The knock-down effect induced by PguCas13b/gPAS was significantly milder than that with PguCas13b/gUSE. (F, G) Schematic view of the single (F) or double (G) gRNA-RNA mismatch positions within gUSE (Left), and the effects (Right) of corresponding mismatches on dPguCas13b PAS perturbation efficiency. Except the one at the very 3′ end, all the other single mismatches significantly impaired the repressive effect of dPguCas13b/gUSE complex. Among these mutants, mismatches at 5′ region of gRNA more significantly attenuated the interference on PAS usage than those at 3′ region (F). A stronger effect could be observed for double mismatches on the gRNA sequence, with a similar positional bias (G). B-G: For each experiment, three independent repeats were performed. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, paired two-way Student's t-test. (H) The usage of endogenous PASs was measured by 3′ end mRNA sequencing in HEK293TAPA reporter cells co-transfected with dPguCas13b and gUSE or gNT. Out of 16,692 expressed PASs, only four PASs showed differential usage (FDR < 0.05 and difference of PAS usage > 0.1) between the cells transfected with gUSE and gNT. (I) The mRNA abundance of endogenous genes was measured by RNA-seq in HEK293TAPA reporter cells co-transfected with dPguCas13b and gUSE or gNT. Out of 14,812 expressed genes, no differential gene expression (P-value < 0.01 and log2 fold change > 1) between the cells transfected with gUSE and gNT was observed. H-I: For 3′ end mRNA sequencing, and RNA-seq, two replicates were performed for each experiment.