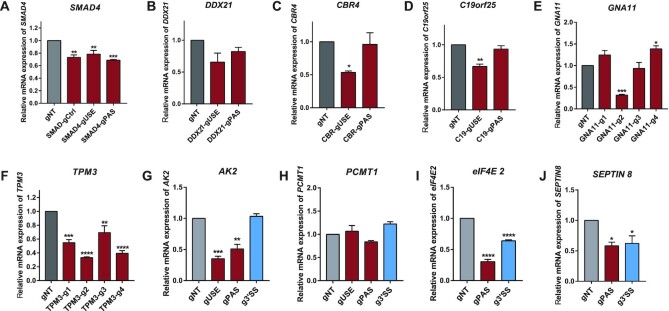
Figure 5.
CRISPR-iPAS targeted at proximal PAS often exerted negative effect on the total mRNA abundance. The total mRNA abundance was measured in HEK293T cells with 3xEGFP-dPguCas13b and indicated gRNAs transfected for gene SMAD4 (A), DDX21 (B), CBR4 (C), C19orf25 (D), GNA11 (E), TPM3 (F), AK2 (G), PCMT1 (H), eIF4E2 (I) and SEPTIN8 (J). For the six genes with tandem 3′UTR APA, including SMAD4, DDX21, CBR4, C19orf25, GNA11 and TPM3, the total mRNA level was often significantly decreased when their proximal PAS was perturbed by CRISPR-iPAS (A–F). The decrease of total mRNA level induced by different gRNAs appeared to correlate with their inhibitory effects on the PAS usage. In the genes containing intronic APA, CRISPR-iPAS targeting the proximal PAS significantly reduced the total mRNA level of AK2 (G), but caused no effect on PCMT1 (H), whereas no effect was observed with CRISPR-IPAS targeting 3′SS on either AK2 or PCMT1 (G, H). For the genes with alternative-terminal-exon APA including SEPTIN8 (I) and eIF4E2 (J), CRISPR-IPAS targeting either PAS or 3′SS significantly decreased the total mRNA abundance.