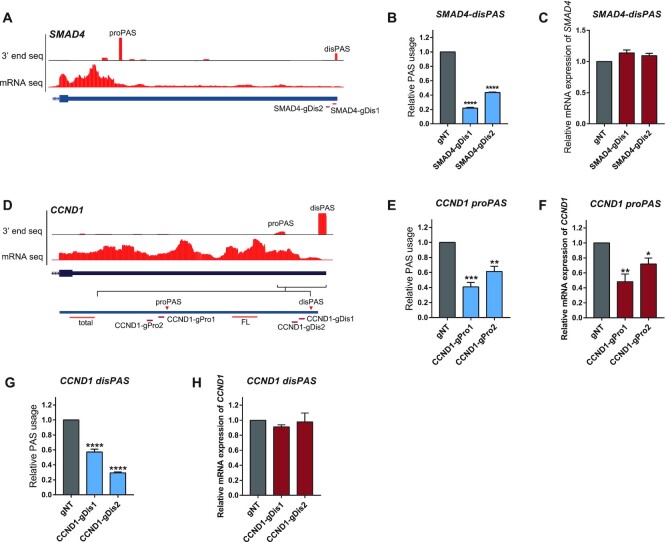
Figure 6.
CRISPR-iPAS targeted at distal PAS exerted no negative effect on the mRNA abundance. (A) UCSC genome browser tracks for RNA-Seq and 3′end mRNA Seq were shown for the last exon of the human gene SMAD4. The position of gRNAs targeting the distal PAS (disPAS) were indicated. (B) The usage of the distal PAS in gene SMAD4 was repressed after transfection of 3xEGFP-dPguCas13b together with the two gRNAs targeting the PAS signal (gPAS). (C) None of the two gRNAs targeting the PAS signal (gPAS) caused significant negative effect on total mRNA abundance of SMAD4. (D) UCSC genome browser tracks for RNA-Seq and 3′end mRNA Seq was shown for the last exon of the human gene Cyclin D1 (CCND1). The proximal PAS (proPAS) and the distal PAS with the dominant usage (disPAS) were indicated. Two gRNAs were designed to target the PAS signal of the distal PAS and proximal PAS, respectively. (E–H) gRNAs targeting either (E) its proPAS or (G) disPAS could significantly repress the usage of their target PAS, whereas the gRNAs targeted at proximal PAS significantly decreased the total mRNA abundance (F), those targeted at distal PAS did not at all (H). For each experiment, three independent repeats were performed. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, paired two-way Student's t-test.