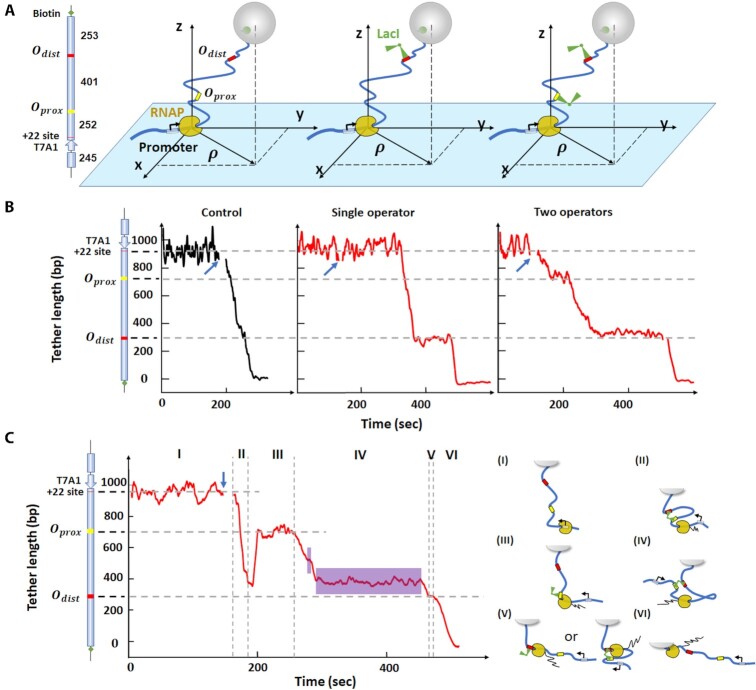
Figure 1.
RNAP can transcribe through a LacI-mediated loop after a pause. (A) The DNA tethers for TPM include DNA (thick, blue line), LacI binding sites (yellow and red) and LacI (green, V-shape). The black arrow at the promoter indicates the direction of transcription. RNAP is stalled at the + 22 site before resuming RNA synthesis in the presence of all NTPs. The cartoons on the right illustrate three scenarios: RNAP elongation without LacI (left), with LacI bound to single operator (middle), or with two LacI bound to two operators separately (right). Each cartoon corresponds to the trace shown below it in Panel B. (B) Representative transcription records. Left: control record without LacI. Center: transcription in the presence of 10 nM LacI on a template with the distal operator only. Right: transcription in the presence of 10 nM LacI and two LacI binding sites. At this concentration, each binding site is likely to be occupied by a different LacI tetramer, so no looping is likely. The blue arrows indicate the time at which NTPs were introduced. (C) Transcription in the presence of 0.2 nM LacI to promote looping. The vertical dash lines identify six intervals (I - VI) in the progress of RNAP along the DNA template and the cartoons on the right depict the likely conformation of the transcription elongation complex in each interval. The purple areas in region IV indicate random pauses between operators. The cartoon on the left of the y-axis in panels B and C shows the features of the DNA template used in TPM measurements. From top to bottom: a T7A1 promoter, a stall site at + 22, a promoter-proximal binding site (Oprox) and promoter-distal binding site (Odist). The horizontal dashed lines indicate the position of the LacI binding site(s) in the construct, and the expected location of pauses in the record.