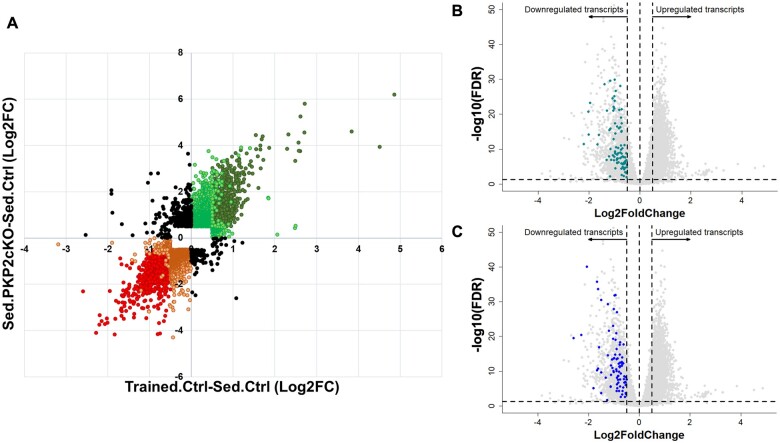
Figure 2.
Additive effects and preserved directionality of transcriptomic changes induced by exercise and by PKP2 knockdown. (A) Cartesian map where each dot represents the log2Fc value of a given transcript in the ‘trained control’ dataset (X-axis; differential transcriptome of Trained.Ctrl-Sed.Ctrl; same dataset as in Figure 1C and D) against the log2FC recorded for the same transcript in the sedentary PKP2cKO dataset (Sed.PKP2cKO-Sed.Ctrl dataset; Y-axis; dataset previously reported in Ref.3). Notice that, as an overall trend, transcriptomic changes caused by training in controls showed the same direction as those caused by loss of PKP2 in sedentary animals. Red circles in bottom left quadrant show transcripts significantly down-regulated (false discovery rate <0.05), with absolute log2FC values ≥0.5, both in the trained control and in the sedentary PKP2cKO dataset. Similarly, dark green circles in upper right quadrant correspond to transcripts significantly up-regulated (false discovery rate <0.05) with absolute log2FC values >0.5 in both datasets. Orange and light green circles show transcripts with absolute log2FC values >0.5 in one dataset but not in the other, regardless of their significance. Orange and light green data points also represent data where Log2FC’s were >0.5 but the value did not reach statistical significance in one or both of the datasets. Black circles denote data points for which the directionality of the change in one dataset was opposite to that of the other. The latter amounted to only 7.2% of the complete dataset. Red and orange data points correspond to 44.3% and 55.7% of the total data points in the 3rd quadrant, respectively. Green and light green data points correspond to 41.3% and 58.7% of the total data points in the 1st quadrant, respectively. (B and C) Grey dots represent differential transcriptome after exercise (same as in Figure 1A). Turquoise (B) and blue dots (C) mark transcripts identified by Montnach et al. 14 as components of the human PKP2 gene network.