Figure 3.
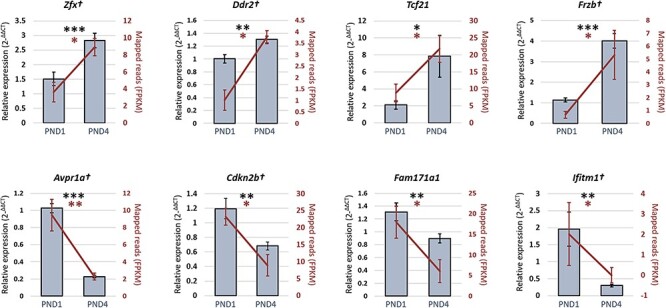
The RT-qPCR validation of differentially expressed genes within neonatal mouse granulosa cells. In verifying the RNA-seq data (represented on the secondary y-axis by the fragment per kilobase of exon per million, FPKM, for each mapped read), eight genes that displayed significantly different levels of expression in PND1 granulosa cells compare to PND4 granulosa cells (see Table 3) were selected for orthogonal validation using RT-qPCR. Validation experiments were performed in triplicate using ≥ six distinct pools of biological samples (n = 4–15 animals per sample), statistical significance was investigated via a student t-test. The 60S ribosomal protein L19 gene was employed as an endogenous control to normalize the expression levels of target genes. Six out of 8 genes remained significant after Bonferroni correction for multiple comparison and are indicated by †. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
