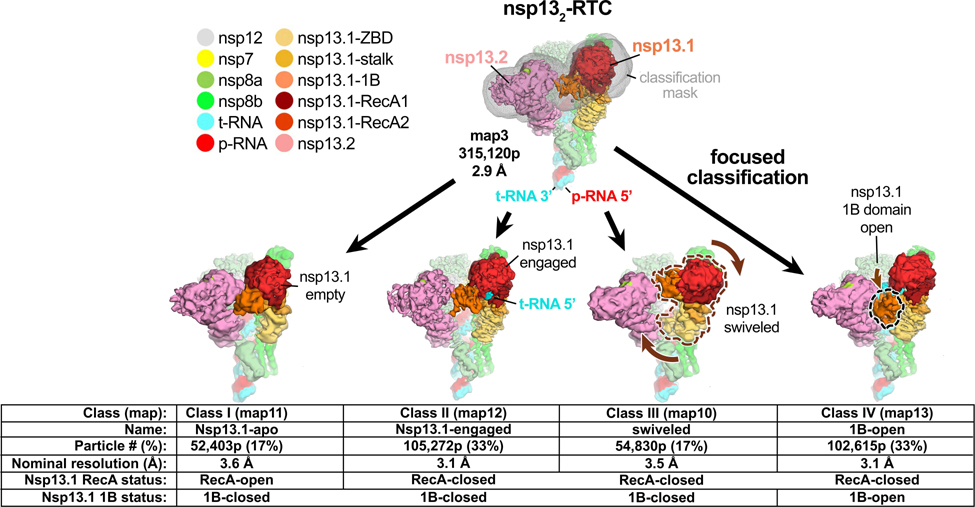
Fig. 1 |. Four conformational states of the nsp132-RTC.
(top) Cryo-EM density (map3, local-resolution filtered) colored according to the code on the left. A mask was constructed surrounding the nsp13T and nsp13F 1B, RecA1, and RecA2 domains (grey mesh). The 315,120 particles were divided into four distinct structures (class I, II, III, and IV) by focused classification inside the mask, followed by further refinement (Extended Data Figs. 2, 6). Class II contained the most particles, and the nsp13T RecA domains were completely closed (Extended Data Fig. 6), entrapping the 5’-t-RNA segment in a groove between the two RecA domains and the 1B domain (Fig. 2). Therefore, class II (nsp13T-engaged) was used as a reference for comparison of the other structures. Each class was characterized by one dominant conformational change: class I) nsp13T-apo, the RecA domains were completely open (Extended Data Fig. 6) and devoid of RNA (Fig. 3), class III) swiveled, the nsp13T protomer as a whole was rotated 38° as shown (Fig. 5), class IV) 1B-open, the nsp13T 1B domain was rotated open by 85° (Fig. 4).
Also see Extended Data Fig. 6 and Supplementary Videos 1 and 2.