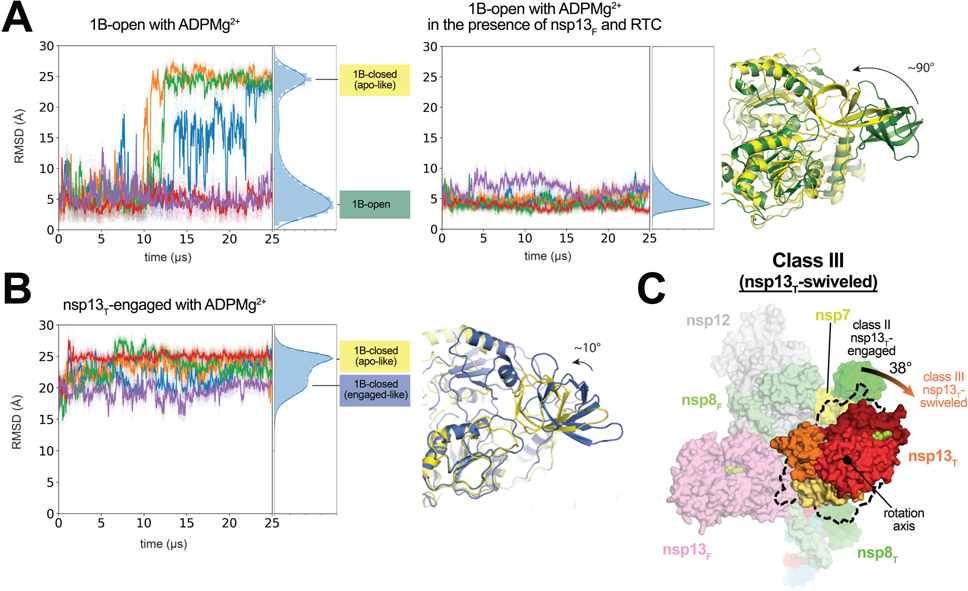
Fig. 5 |. In the nsp13T-swiveled structure, the entire nsp13T promoter is rotated.
A. 10 independent simulations of ADPMg2+-bound nsp13T, starting from the 1B-open cryo-EM structure, in isolation (five simulations, left-hand plot) and as part of the nsp132-RTC complex (five simulations, right-hand plot). Values plotted represent the heavy-atom rmsd of the 1B domain (nsp13 residues 150–228) compared to the 1B domain in the 1B-open cryo-EM structure (aligned on the RecA1 domain). The rmsd histograms on the right of each plot represent aggregate values across all five simulations. Representative structures of the two major conformations from simulations are shown (right, colored according to the histogram labels).
B. Five independent simulations of ADPMg2+-bound nsp13T, starting from the nsp13T-engaged state. Values plotted represent the heavy-atom rmsd of the 1B domain compared to the 1B domain in the 1B-open cryo-EM state (aligned on the RecA1 domain). The rmsd histograms on the far right represent aggregate values across all five simulations. Representative structures of the two major conformations from the rmsd histogram from simulations are shown (right).
C. Front view of the nsp13T-swiveled structure, highlighting nsp13T. The position of the nsp13T promoter in the nsp13T-engaged structure is illustrated by the dashed black outline. The nsp13T protomer of the nsp13T-swiveled structure is rotated by 38° as shown.
See also Extended Data Fig. 7.