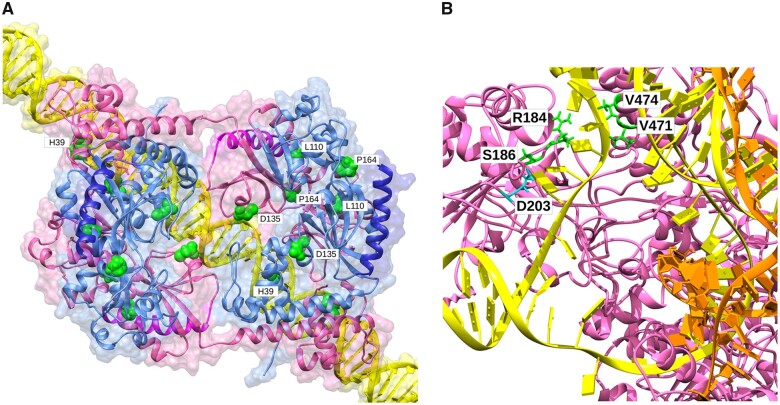
Fig. 7.
Modeling of the effect of AR and compensatory mutations in the structures of AmpR and RpoB. (A) Proposed model of the AmpR tetramer bound to the putative promoter of ampC of P. aeruginosa PA14. Mutated amino acids are shown as balls for better identification. Only amino acids in one pair of monomers have been labeled. The C-terminal regions affected by 274 frame shifts are highlighted. D135N and H39Y would affect promoter recognition, F19fs truncates the protein, L110P would destabilize the complex, P164L would destabilize the effector binding pocket, and G274fs would affect interaction with RNAP. (B) Proposed model of RpoB bound to a DNA bubble and a nascent RNA molecule. Mutated amino acid D203, whose mutation causes the resistant phenotype (possibly through a change in promoter specificity), and amino acids with reversion mutations are shown as sticks. D203E would affect the interaction with the open DNA bubble, pushing the nontranscribed (NT) strand toward the channel between R184 and V471-V474. Reversion mutants S186F, R184C, V471E, and V474E would correct stacking, tensions, and “scrunching” distortions in the NT strand.