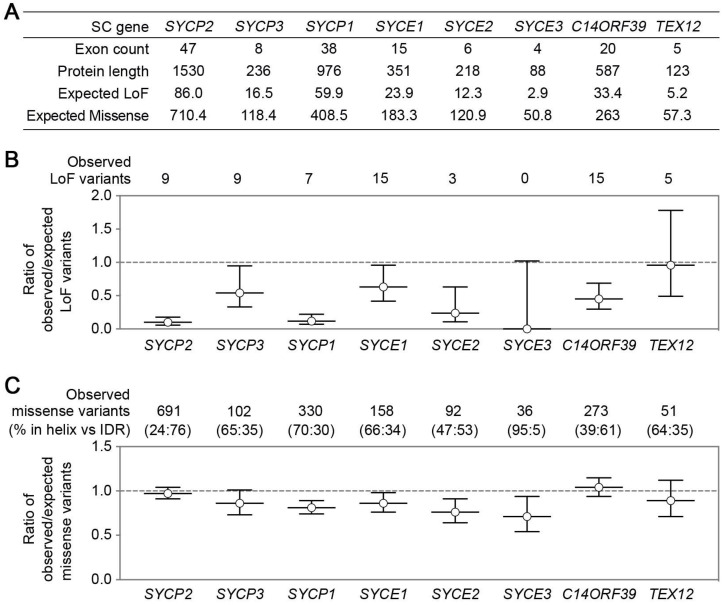
Figure 2.
SC gene variants in the general population. SC gene variants data was collected from the gnomAD database 69. (A) A table summarizes SC gene exon counts, protein product lengths, and the expected number of loss-of-function (LoF) or missense variants. (B) The ratio of the observed/expected number of LoF variants in SC genes. Error bars represent a 90% confidence interval. The numbers of observed LoF variants are indicated on the top. (C) The ratio of the observed/expected number of missense variants in SC genes. Error bars represent a 90% confidence interval. The numbers of observed missense variants and their distribution in α-helix or IDR regions (shown as percentages in parentheses) are indicated on the top. The expected variant numbers and ratio of observed/expected variants with confidence intervals were determined with a depth corrected mutation probability reported in 89.