Abstract
The genus Silene brings many opportunities for the study of various processes involved in the evolution of dioecy and young sex chromosomes. Here we focus on a dioecious clade in Silene subgenus Silene and closely related species. This study provides improved support for monophyly of this clade (based on inclusion of further dioecious species) and a new estimate of its age (ca 2.3 million years). We observed a rise in adaptive evolution in the autosomal and pseudoautosomal parts of the genome on the branch where dioecy originated. This increase is not a result of the accumulation of sexually antagonistic genes in the pseudoautosomal region. It is also not caused by the coevolution of genes acting in mitochondria (despite the possibility that dioecy along this branch could have evolved from a nucleo-cytoplasmic male sterility-based system). After considering other possibilities, the most parsimonious explanation for the increase seen in the number of positively selected codons is the adaptive evolution of genes involved in the adaptation of the autosomal part of the genome to dioecy, as described in Charnov's sex-allocation theory. As the observed coincidence cannot prove causality, studies in other dioecious clades are necessary to allow the formation of general conclusions.
This article is part of the theme issue ‘Sex determination and sex chromosome evolution in land plants’.
Keywords: genome evolution, dioecy, autosome evolution, sex chromosome, Silene
1. Introduction
Separate sexes are generally associated with animals but they also occur in plants [1]. Once a sex-determining gene appears in the genome, many evolutionary processes are initiated on the sex chromosomes [2]. Moreover, many autosomal genes are expressed in a sex-specific manner, and their expression is regulated by the presence of sex-determining genes [3,4]. These sex-specific differences in gene expression lead to the formation of male and female individuals. In addition, because males invest more in producing a greater number of offspring, whereas females prioritize quality [5], the presence of separate sexes could result in the adaptation of autosomal genes for a plastic response to these different breeding strategies. Thus, the autosomal genes could be selected to function in both types of sex-specific expression patterns. In this case, the adaptation should be visible as a significant increase of the number of adaptively evolved genes on a phylogenetic branch leading to species with separate sexes.
Many plant genera contain recently evolved dioecious clades. An example is found in the genus Silene. Dioecious Silene species are well-suited to this study due to the availability of closely related non-dioecious species, transcriptomic data for both the dioecious species and their non-dioecious relatives, and genetic maps (reviewed in [6]). In Silene, apart from dosage compensation studies [7,8], adaptation of the transcriptome to the dioecy has also been studied from a quantitative point of view [9]. The results [9] showed that in Silene latifolia many differences in gene expression (when compared to gynodioecious relatives) appear connected with the adaptive changes beneficial to S. latifolia females. These changes in gene expression occurred not only in X-linked but also in autosomal sequences. Muyle et al. [10] used a population genomics approach to study the influence of dioecy on genetic diversity and selection efficacy. This approach showed that neither genetic diversity nor selection efficiency is negatively influenced by dioecy and that the dioecious Silene species do not show an increased mutation load. However, it is not known whether the presence of dioecy and the corresponding separation of male and female reproductive programmes affect the evolution of the coding regions in the autosomal part of the genome.
2. Material and methods
To ascertain the position of Silene sibirica in the phylogeny of Silene subgenus Silene, a dataset from Slancarova et al. [11] was used together with publicly available sequences and our sequences (electronic supplementary material, table S1). Here we use subgenus Silene as defined by Jafari et al. [12]. Phylogenetic analysis was done using the IQ-TREE 2 [13] on the alignment file obtained by concatenation of alignments for each sequence (12 640 bp). The dataset was analysed using separate models for each sequence (partition) using the maximum-likelihood method [14]. The appropriate model for each partition was found using ModelFinder [15]. The branch support was computed using the SH-like approximate likelihood ratio test [16] and non-parametric bootstrap with 500 replicates. To check for long-branch attraction, the phylogenetic analysis was also performed solely with the species of Silene subgenus Silene. Phylogenetic analysis performed using MrBayes v. 3.2.6 [17–19] was run for ten million iterations using the same partitioned dataset as in the case of IQ-TREE. The dataset including MrBayes block is available as electronic supplementary material, dataset S1. Plant breeding systems in subgenus Silene were marked according to [20]. The ancestral states were analysed using the maximum-likelihood variant of the MultiState method in BayesTraits [21], using the tree presented in figure 1. The Bayesian analysis of ancestral states was performed using the Bayesian variant of MultiState method (reversible jump MCMC, gamma prior and 100 million iterations) in BayesTraits, using a set of trees prepared in MrBayes to construct the tree presented in electronic supplementary material, figure S1. These analyses focused on three nodes: the most recent common ancestor (MRCA) of dioecious clade of subgenus Silene, the MRCA of this dioecious clade and S. sibirica; and the MRCA of S. sibirica, Silene paradoxa and the dioecious species. At these nodes, probabilities of three possible states were evaluated: non-dioecious species (hermaphrodite and gynodioecious species), subdioecious species and dioecious species.
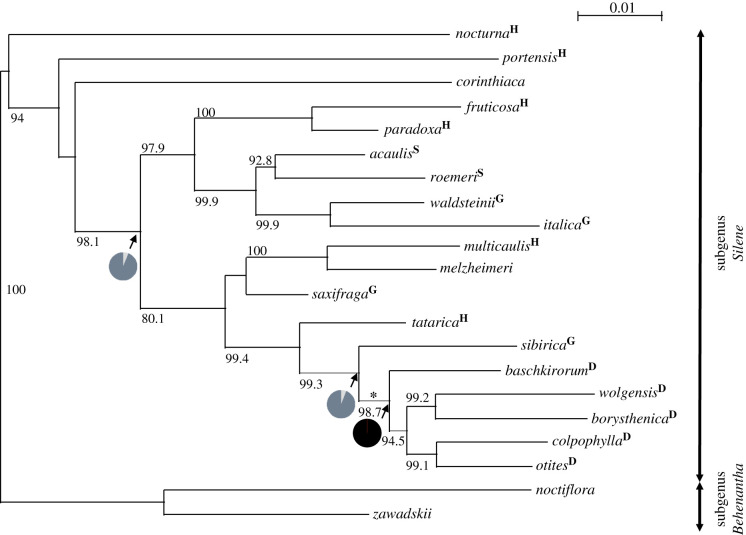
Figure 1.
Phylogenetic tree of the dioecious clade of subgenus Silene and related species. The tree was constructed by the maximum-likelihood method performed using IQ-TREE 2 [13]. The values indicated at the nodes are SH-aLRT (Shimodaira–Hasegawa approximate likelihood ratio test) supports (shown for values of 0.80 or greater). Dioecious species are marked with a ‘D’, gynodioecious species with a ‘G’, hermaphroditic species with an ‘H’ and the subdioecious species are indicated with an ‘S’. Missing symbol means that the species is either hermaphrodite or gynodioecious but their breeding system was not studied in detail. The branch where dioecy and sex chromosomes evolved is marked by an asterisk. The pie charts show the probabilities of the ancestral states in three nodes as obtained in BayesTraits. Dioecy is marked by black colour, hermaphroditism and gynodiecy are marked in dark grey and subdioecy is marked by light grey.
For the analyses of omega (ω) estimates, the RNAseq dataset from the previous work [6] was supplemented with samples from Silene nocturna (SRR6040876; our sequencing; NextSeq 500 (paired-end reads 2 × 75)) and S. paradoxa (SRR999298-SRR999299, publicly available). Basic statistics describing these samples and other samples for which this information was not available is presented in electronic supplementary material, table S2. Silene pseudotites has been excluded from the dataset based on its suspected hybrid origin. The methods used for the preparation of the previous dataset and details concerning basic statistics for the majority of the sequencing data are described in [6]. For most species, the phasing procedure was based on read-based phasing of one individual; only in Silene otites, Silene colpophylla and Silene borysthenica was pedigree-based phasing applied. Pedigree-based phasing showed the phased genotypes present in the parents and several offspring. One allele was again randomly chosen. The orthogroups showing more than two alleles in some species were excluded from the dataset. The reads of S. nocturna and S. paradoxa were separately assembled using Trinity [22]. The obtained assemblies were treated with an Evigene pipeline to reduce duplications and used as a reference. Mapping was done using Bowtie2 -- very-sensitive-local option [23], the SNPs were called via FreeBayes [24] and phasing was performed with WhatsHap [25]. The regions with coverage lower than 2 were masked using the maskfasta method in BEDTools [26]. Masking was done on a sample-by-sample basis. The phased sequences were added to the original alignments with the MAFFT aligner [27] based on the best reciprocal blast hit search [28]. On average, 1.24% positions were missing per species. dS (synonymous substitution rate) estimated between S. sibirica and dioecious species were subsequently compared to further identify potential paralogs. The phylogenetic tree was constructed using both phased alleles. Phylogenetic analysis was done using the StarBeast2 module in BEAST v. 2 [29,30]. Two independent chains were run for 1000 000 000 states (Yule model, birth–death). For the dating, the calibration of the MRCA of S. nocturna and the other species was done similarly to Balounova et al. [6] based on estimated divergence per synonymous site dS (substitutions per synonymous site) and the divergence time estimates for several angiosperm lineages (Brassicaceae, Malvaceae, Euphorbiaceae, Fabaceae, Cucurbitaceae, Rosaceae, Solanaceae and Poaceae), so as not to depend on a single fossil record or phylogenetic tree position, which yielded a mean substitution rate of 5.35 × 10−9 synonymous substitutions/site/year [31]. dS values for the distance of individual species of the dioecious clade of subgenus Silene to S. nocturna were estimated using KaKs_Calculator 2.0 program [32], and the mean value was used for the calibration of the tree. The resulting dataset (electronic supplementary material, dataset S2) did not contain any of the completely sex-linked genes identified previously [6,8].
For the PAML (phylogenetic analysis by maximum likelihood) analyses, only one allele per gene was randomly chosen. PAML analyses were used to determine whether some expressed sequence tags (ESTs) evolved under selective pressure. The CODEML program of PAML [33] was used to estimate the ratio (ω) of the non-synonymous substitution rate (dN) to the synonymous substitution rate (dS). As the reference tree, the phylogenetic tree constructed as described above was used. The equilibrium frequencies of codons were calculated from the nucleotide frequencies (CodonFreq = 2) because it best fits the data as calculated by second-order AIC. All models in CODEML were run at five different initial ω values (ω = 0.1, 0.2, 0.5, 1, 2), and no problems with the convergence were observed. Both branch and branch-site models were applied to the branches preceding, including and following the origin of dioecy. In the branch analyses (model = 2, NSsites = 0) two-ratios models were compared to three-ratios models to reveal whether the ω values differed significantly. The resulting log-likelihood values were evaluated using likelihood-ratio tests to determine any statistical significance of the difference. The chi2 program of PAML was used to estimate the p-values. The confidence intervals for proportion were computed online (http://vassarstats.net/prop1.html) using the method with continuity correction [34] that is derived from a procedure outlined in [35]. The results obtained in CODEML were further confirmed using a test for relaxed selection in the RELAX program [36] of the HyPhy package [37,38]. Because the results did not converge well, the analysis was run 500 times and the results with the highest log-likelihood were used.
The modified model A (model = 2, NSsites = 2) of the branch-site models was applied to calculate the percentages of purifying, neutral and adaptive codons in the analysed ESTs, the ω values, the Bayes Empirical Bayes probability of codons being either purifying, neutral or adaptive and to identify ESTs with adaptively evolved codons. The signed-rank Wilcoxon test was used to compare the ω values of the codons with ω less than 1 between the branches before and during the origin of dioecy. The chi-squared goodness-of-fit test was used to calculate the p-value of adaptively evolving codons that arise from neutrally evolving codons. The Pearson's chi-squared test was used to calculate the p-value corresponding to a random presence of genes acting in mitochondria among the genes with positively selected codons.
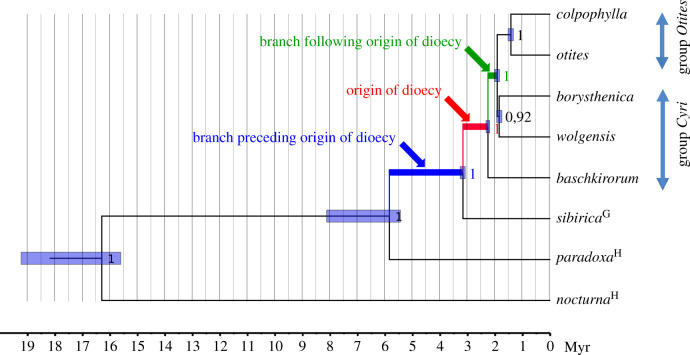
The analyses performed in PAML and HyPhy packages were based on the comparison of the ω values between branches on the phylogenetic tree shown in figure 2. The ω value of the branch on which most likely dioecy (and sex chromosomes) originated (marked by an asterisk in figure 1 and highlighted in red in figure 2) was compared with the ω value of the preceding branch (blue in figure 2) and with the ω value of the following branch (green in figure 2). Although there are two branches following the origin of dioecy: one leading to S. baschkirorum, the other leading to the other dioecious species of subgenus Silene, only the latter was used in the analyses as studies of ω are not recommended in evolutionarily young lineages [39].
Figure 2.
Phylogenetic tree of the dioecious clade of subgenus Silene and closely related species. Gynodioecious species is marked with a ‘G’ and hermaphroditic species with an ‘H’. The species belonging to groups Cyri and Otites are dioecious. The colours of the highlighted branches refer to CODEML and RELAX analyses. For details, see §3.
3. Results and discussion
In this study, we used the currently available sequencing data obtained for the dioecious clade of Silene subgenus Silene (as defined by Jafari et al. [12]) to investigate the possible correlation between the rise of dioecy and the evolution of non-sex-linked sequences. This dioecious clade has previously been grouped together based on morphological traits [40]. Later molecular studies have supported this view but only included a small number of species [6,11,12]. As the monophyly of this dioecious clade is an important prerequisite for our study, we performed phylogenetic analyses with a better representation of these dioecious species (see electronic supplementary material, table S3). The studied clade includes 15 species, all of them reported as dioecious (see electronic supplementary material, table S3). According to Wrigley [40], the centre of distribution of these species is the Ukraine and South Russia, but particular species are spread over almost all of Europe and a big part of Asia (see also electronic supplementary material, figure S2). The phylogenetic analyses show that the eleven dioecious species studied by us form a strongly supported monophyletic clade (SH-aLRT = 99.6, nonparametric bootstrap support = 95, Bayesian posterior probability = 1.00); see electronic supplementary material, figure S1.
Previous results [6] have shown that the gynodioecious species S. sibirica appears to be a close relative to the dioecious clade mentioned and our analyses support these findings (figure 1 and electronic supplementary material, figure S1). Analysis of the ancestral states of the breeding system in genus Silene [11], along with our own findings, suggest that the MRCA of the dioecious clade was probably dioecious (greater than 99% probability). Moreover, in all genetically studied species of this clade (S. otites, S. colpophylla, S. borysthenica and S. pseudotites), sex chromosomes have been found [6,8]. The other species in subgenus Silene (with the exception of Silene roemeri and Silene acaulis) are gynodioecious or hermaphroditic, and the analysis of the ancestral states of the breeding system [11] and our results obtained in BayesTraits [21] support the view that the ancestral species preceding MRCA of this clade was most likely gynodioecious or hermaphroditic. Thus, the transition from gynodioecy or from hermaphroditism to dioecy most likely occurred on the branch marked by an asterisk in figure 1 and highlighted in red in figure 2.
The topology in figure 2 is mostly in accordance with previous topologies that did not include S. nocturna and S. paradoxa. A minor difference is that recent results support the clade joining two members (S. borysthenica and Silene wolgensis) of the group Cyri (defined previously according to phenotypic markers [38]), while in our past analyses, the group Cyri appeared as completely polyphyletic [6]. The newly obtained dating, based on the estimation of the synonymous divergence of the outgroup (S. nocturna) from the other species, suggests the age of the studied dioecious clade to be approximately 2.3 million years, which is in the range of the previous estimate based on calibration by fossils (1.17–2.60 million years) [11].
The ω ratio (ω = dN/dS) is one of the most widely used statistical tests applied to quantify selection pressures acting on protein-coding regions. This measure compares the rate of substitutions at silent sites (dS), which are presumed neutral, to the rate of substitutions at non-silent sites (dN), which possibly experience selection. The comparison of the two- and three-ratio branch models in the CODEML program of the PAML package [33] shows that the ω value of the branch preceding the origin of dioecy (ω = 0.16; blue in figure 2) is lower than the ω value of the branch where dioecy originated (ω = 0.29; red in figure 2). This difference is highly significant (p < 10−99, likelihood ratio test; LRT). After the formation of the dioecy (green in figure 2), the ω value significantly decreased (ω = 0.26; p < 10−99, LRT).
Because the increase of the ω values can be due to the changes in the number of either positively or neutrally selected sites, we compared branch-site models (table 1). The branch where dioecy originated showed a significant percentage of positively selected codons (p < 10−99, LRT) therefore increasing the overall ω value present in the branch where dioecy originated. On the other hand, we did not detect codons under positive selection in the branch preceding dioecy (blue in figure 2) (p > 0.99, LRT). Moreover, the ω values of sites under purifying selection did not increase significantly (p = 0.95, Wilcoxon test). To confirm these results, we performed tests for relaxed selection using the RELAX program [36] of the HyPhy package [37,38]. During the transition from gynodioecy to dioecy, selection intensification (p = 0.00; K = 1.55) was observed. On the other hand, when the branch including the origin of dioecy and the branch after the origin of dioecy were compared, non-significant relaxation was detected (p = 0.339, K = 0.91). Most of the identified adaptively evolved codons (63 out of 87) are recruited from the neutrally evolved codons, which is significantly more than by chance (p < 10−16, chi-squared goodness-of-fit test).
Table 1.
Branch-site analysis of Silene transcriptome before and after sex chromosome evolution. Confidence intervals (CI) are given for the differences in percentage between the neighbouring rows. Statistically significant values are highlighted in bold.
|
ω < 1 |
ω = 1 |
ω > 1 |
||||
|---|---|---|---|---|---|---|
| branch | percentage (%) | 95% CI of percentage difference (%) | percentage (%) | 95% CI of percentage difference (%) | percentage (%) | 95% CI of percentage difference (%) |
| preceding sex chromosome evolution | 80.496 | 19.504 | 0 | |||
| −0.19, 0.31 | −0.17, 0.34 | 0.02, 0.03 | ||||
| including sex chromosome evolution | 80.557 | 19.419 | 0.024 | |||
| −0.2, 0.31 | −0.17, 0.32 | 0.02, 0.03 | ||||
| following sex chromosome evolution | 80.497 | 19.503 | 0 | |||
The increase in the number of positively selected codons on the branch including the origin of dioecy (and sex chromosomes) cannot be caused by the selection of sexually antagonistic genes on the sex chromosomes [41], because the analysed dataset does not contain any completely sex-linked genes. Moreover, it cannot be the result of the accumulation of sexually antagonistic genes in the pseudoautosomal region, as only one of the studied ESTs was located in this part of the sex chromosomes.
The dioecy in the studied clade most likely evolved from gynodioecy as there are no monoecious species in genus Silene ([20]; see also figure 1). Because several cases of the nucleo-cytoplasmic type of gynodioecy are known from Silene [42–46], the genetic fixation of a male-sterile cytoplasm and subsequent recruitment of a fertility restorer as the sex-determining gene is possible [47]. The fixation of a certain type of male-sterility-causing cytoplasm could influence the evolution of the nuclear genes involved in mitochondrial metabolism. However, among the 42 ESTs identified as having at least one adaptively evolved codon on the branch including the origin of dioecy, we did not find an overrepresentation of proteins located in the mitochondria (7 ESTs; p = 0.77, Pearson's chi-squared test).
The observed increase in the number of positively selected codons could have been caused by the selection of genes involved in the adaptation of the genome to dioecy as described in the sex-allocation theory [5], which states that female plants allocate more resources to the quality of seeds whereas male plants allocate resources into the quantity of pollen. However, based on phenotypic data, the adaptation of the genome to the dioecy appears complex and dependent upon a wide variety of genes [48]. Indeed, the genes identified as adaptively evolving on the branch where dioecy evolved (see electronic supplementary material, dataset S3) show diverse characteristics. Detailed studies of phenotypic sexual dimorphism in dioecious clade of subgenus Silene have not yet been done but as reported already by Correns [49], the males of S. otites show habitus that gains the attention of observers more readily than that of females, possibly leading to the overestimation of male bias often reported in this species (reviewed by [50]). In S. otites, male plants have been reported to be more sensitive to drought stress and to be flowering earlier in the season [50]. The coincidence between the increased number of adaptively evolved codons in autosomes and the evolution of separate sexes documented in this study deserves to be followed up in future work in other recently evolved dioecious species.
Acknowledgements
We would like to thank Prof. Susanne Renner (Department of Biology, Washington University, Saint Louis, MO, USA) and Dr Jiri Danihelka (Department of Botany and Zoology, Faculty of Science, Masaryk University, Brno, Czech Republic) for their comments. We would like also to thank Dr Jiri Danihelka and Prof. Aleksandr L. Ebel (Department of Botany, Tomsk State University, Russia) for plant material.
Contributor Information
Jitka Zluvova, Email: jitka@ibp.cz.
Bohuslav Janousek, Email: janousek@ibp.cz.
Ethics
This work is in silico, with no ethics requirements.
Data accessibility
The data generated in this project have been deposited in Genbank. For accession numbers see electronic supplementary material, tables S1 and S2.
Authors' contributions
J.Z.: Conceptualization, formal analysis, investigation, methodology, project administration, software, supervision, validation, visualization, writing—original draft, writing—review and editing; Z.K.: funding acquisition, project administration, writing—review and editing; R.H.: project administration, resources, writing—review and editing; B.J.: data curation, formal analysis, investigation, methodology, project administration, software, supervision, validation, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare no competing interests.
Funding
This research was supported by the Czech Science Foundation (Grantová agentura České republiky) grant no. 19-15609S. Computational resources were supplied by the project ‘e-Infrastruktura CZ’ (e-INFRA CZ LM2018140) supported by the Ministry of Education, Youth and Sports of the Czech Republic (Ministerstvo školství, mládeže a tělovýchovy) and by the ELIXIR-CZ project (LM2018131), part of the international ELIXIR infrastructure.
References
- 1.Renner SS, Müller NA. 2021. Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat. Plants 7, 392-402. ( 10.1038/s41477-021-00884-3) [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth D. 2019. Young sex chromosomes in plants and animals. New Phytol. 224, 1095-1107. ( 10.1111/nph.16002) [DOI] [PubMed] [Google Scholar]
- 3.Deegan DF, Engel N. 2019. Sexual dimorphism in the age of genomics: how, when, where. Front. Cell. Dev. Biol. 7, 186. ( 10.3389/fcell.2019.00186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Zhang X, Fatima M, Ma X, Fang H, Yan H, Ming R. 2020. DNA methylome and transcriptome landscapes revealed differential characteristics of dioecious flowers in papaya. Hortic. Res. 7, 81. ( 10.1038/s41438-020-0298-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charnov EL. 1982. The theory of sex allocation. Monogr. Popul. Biol. 18, 1-355. ( 10.1515/9780691210056) [DOI] [PubMed] [Google Scholar]
- 6.Balounova V, et al. 2019. Evolution of sex determination and heterogamety changes in section Otites of the genus Silene. Sci. Rep. 9, 1045. ( 10.1038/s41598-018-37412-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muyle A, et al. 2018. Genomic imprinting mediates dosage compensation in a young plant XY system. Nat. Plants 4, 677-680. ( 10.1038/s41477-018-0221-y) [DOI] [PubMed] [Google Scholar]
- 8.Martin H, Carpentier F, Gallina S, Godé C, Schmitt E, Muyle A, Marais GAB, Touzet P. 2019. Evolution of young sex chromosomes in two dioecious sister plant species with distinct sex determination systems. Genome Biol. Evol. 11, 350-361. ( 10.1093/gbe/evz001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemp N, Widmer A, Charlesworth D. 2018. Has adaptation occurred in males and females since separate sexes evolved in the plant Silene latifolia? Proc. R. Soc. B 285, 20172824. ( 10.1098/rspb.2017.2824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muyle A, et al. 2021. Dioecy is associated with high genetic diversity and adaptation rates in the plant genus Silene. Mol. Biol. Evol. 38, 805-818. ( 10.1093/molbev/msaa229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slancarova V, et al. 2013. Evolution of sex determination systems with heterogametic males and females in Silene. Evolution 67, 3669-3677. ( 10.1111/evo.12223) [DOI] [PubMed] [Google Scholar]
- 12.Jafari F, Zarre F, Gholipour A, Eggens F, Rabeler RK, Oxelman B. 2020. A new taxonomic backbone for the infrageneric classification of the species-rich genus Silene (Caryophyllaceae). Taxon 69, 337-368. ( 10.1002/tax.12230) [DOI] [Google Scholar]
- 13.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530-1534. ( 10.1093/molbev/msaa015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997-1008. ( 10.1093/sysbio/syw037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587-589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307-321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 17.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754-755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 18.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. ( 10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539-542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casimiro-Soriguer I, Buide ML, Narbona E. 2015. Diversity of sexual systems within different lineages of the genus Silene. AoB Plants 7, plv037. ( 10.1093/aobpla/plv037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673-684. ( 10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- 22.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494-1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E, Marth G. 2012Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907. [Google Scholar]
- 25.Patterson M, Marschall T, Pisanti N, van Iersel L, Stougie L, Klau GW, Schönhuth A. 2015. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 22, 498-509. ( 10.1089/cmb.2014.0157) [DOI] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. ( 10.1093/bioinformatics/btq033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinf. 10, 421. ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537. ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogilvie HA, Bouckaert RR, Drummond AJ. 2017. Starbeast2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 34, 2101-2114. ( 10.1093/molbev/msx126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De La Torre AR, Li Z, Van de Peer Y, Ingvarsson PK. 2017. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol. Biol. Evol. 34, 1363-1377. ( 10.1093/molbev/msx069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 8, 77-80. ( 10.1016/S1672-0229(10)60008-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586-1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 34.Newcombe RG. 1998. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat. Med. 17, 857-872. () [DOI] [PubMed] [Google Scholar]
- 35.Wilson EB. 1927. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 22, 209-212. ( 10.1080/01621459.1927.10502953) [DOI] [Google Scholar]
- 36.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820-832. ( 10.1093/molbev/msu400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676-679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
- 38.Pond SLK, et al. 2020. HyPhy 2.5—A customizable platform for evolutionary hypothesis testing using phylogenies. Mol. Biol. Evol. 37, 295-299. ( 10.1093/molbev/msz197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mugal CF, Wolf JB, Kaj I. 2014. Why time matters: codon evolution and the temporal dynamics of dN/dS. Mol. Biol. Evol. 31, 212-231. ( 10.1093/molbev/mst192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrigley F. 1986. Taxonomy and chorology of Silene section Otites (Caryophyllaceae). Ann. Bot. Fenn 23, 69-81. [Google Scholar]
- 41.Rice WR. 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381, 232-234. ( 10.1038/381232a0) [DOI] [PubMed] [Google Scholar]
- 42.Charlesworth D, Laporte V. 1998. The male-sterility polymorphism of Silene vulgaris: analysis of genetic data from two populations and comparison with Thymus vulgaris. Genetics 150, 1267-1282. ( 10.1093/genetics/150.3.1267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lahiani E, Dufaÿ M, Castric V, Le Cadre S, Charlesworth D, Van Rossum F, Touzet P. 2013. Disentangling the effects of mating systems and mutation rates on cytoplasmic [correction of cytoplamic] diversity in gynodioecious Silene nutans and dioecious Silene otites. Heredity 111, 157-164. ( 10.1038/hdy.2013.32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Touzet P, Delph L. 2009. The effect of breeding system on polymorphism in mitochondrial genes of Silene. Genetics 181, 631-644. ( 10.1534/genetics.108.092411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Štorchová H, Stone JD, Sloan DB, Abeyawardana OAJ, Müller K, Walterová J, Pažoutová M. 2018. Homologous recombination changes the context of cytochrome b transcription in the mitochondrial genome of Silene vulgaris KRA. BMC Genom. 19, 874. ( 10.1186/s12864-018-5254-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garraud C, Brachi B, Dufay M, Touzet P, Shykoff JA. 2011. Genetic determination of male sterility in gynodioecious Silene nutans. Heredity 106, 757-764. ( 10.1038/hdy.2010.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zluvova J, Lengerova M, Markova M, Hobza R, Nicolas M, Vyskot B, Charlesworth D, Negrutiu I, Janousek B. 2005. The inter-specific hybrid Silene latifolia x S. viscosa reveals early events of sex chromosome evolution. Evol. Dev. 7, 327-336. ( 10.1111/j.1525-142X.2005.05038.x) [DOI] [PubMed] [Google Scholar]
- 48.Geber MA, Dawson TE, Delph LF (eds). 1999. Gender and sexual dimorphism in flowering plants. Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- 49.Correns C. 1928. Bestimmung, Vererbung und Verteilung des Geschlechtes bei den hoheren Pflanzen. Handbuch der Vererbungswissenschaft, vol. 2, pp. 1-138. Berlin, Germany: Gebruder Borntraeger. [Google Scholar]
- 50.Soldaat LL, Lorenz H, Trefflich A. 2000. The effect of drought stress on the sex ratio variation of Silene otites. Folia Geobot. 35, 203-210. ( 10.1007/BF02803098) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in this project have been deposited in Genbank. For accession numbers see electronic supplementary material, tables S1 and S2.