Figure 1. Binding of apo-specific Mb to RAS.
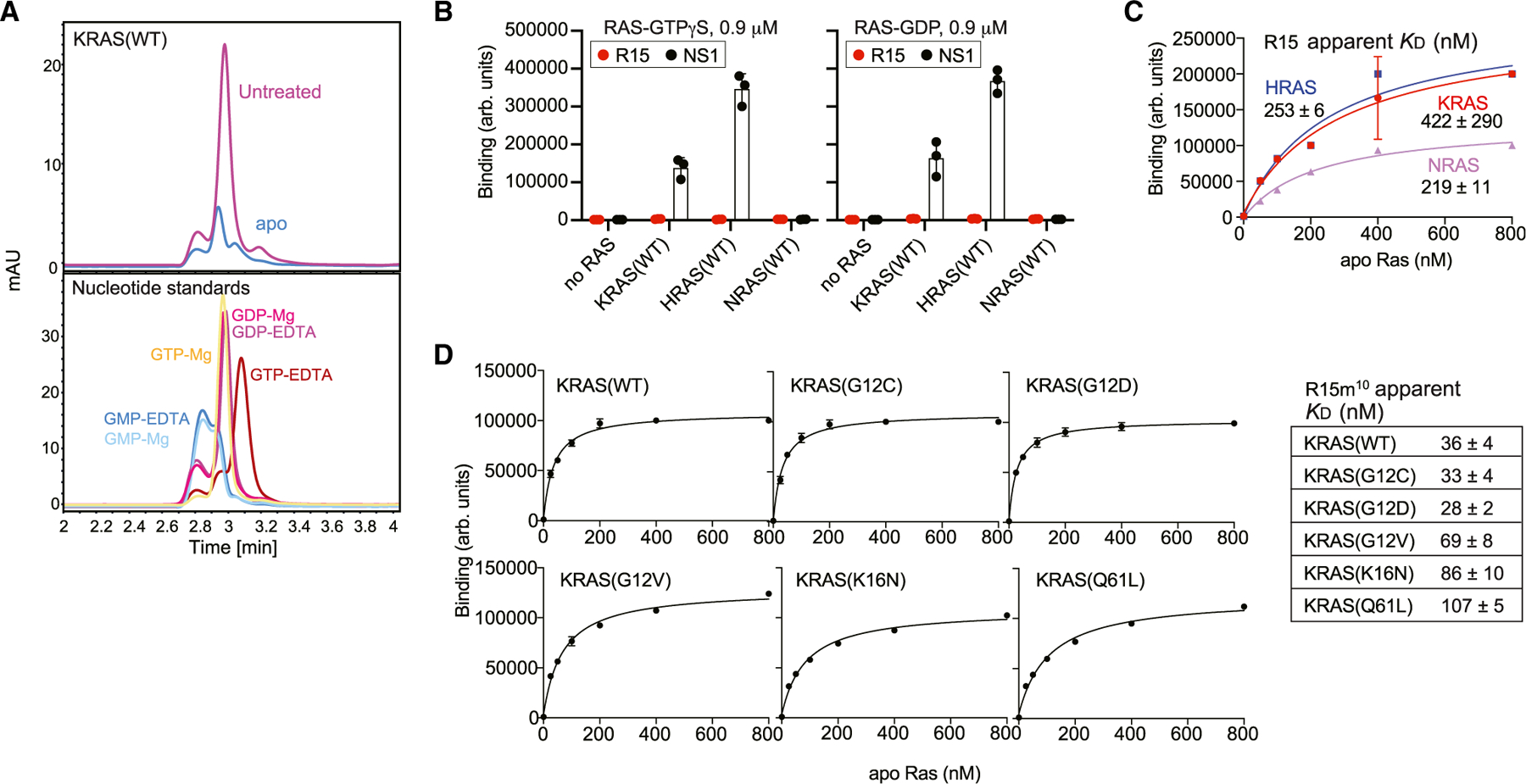
(A) Confirmation of the nucleotide-free nature of apo RAS preparations (Jeganathan et al., 2018). Chromatograms showing nucleotides released from RAS samples (top) and free nucleotide standards in the presence of 0.1 mM EDTA or 0.5 mM MgCl2.
(B) Binding of R15 Mb to GTP- or GDP-loaded RAS and to apo RAS, as measured using yeast surface display. NS1 Mb, which binds to GTP- and GDP-bound states of HRAS and KRAS, was used as a positive control.
(C) Binding titration of R15 Mb expressed on yeast cell surface to nucleotide-free RAS isoforms using flow cytometry is shown. Apparent KD values are shown (mean and SD; n = 3; technical replicates).
(D) Binding titration of R15m10 to various apo KRAS proteins. The table shows apparent KD values (mean and SD; n = 3; technical replicates).
See also Figure S1 for nucleotide-state specificity of R15m10.
