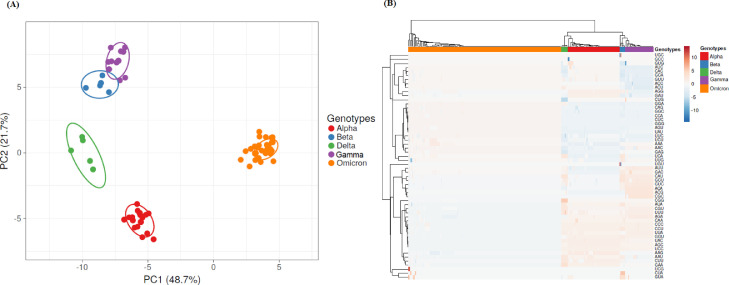
Fig. 1.
PCA of codon usage in Spike proteins from SARS-CoV-2 VOCs strains. In (A) the position of the Spike proteins in the plane conformed by the first two major components of PCA is shown. SVD was used to calculate principal components and unit variance was applied. The proportion of variance explained by each axis is shown between parentheses. Prediction ellipses are such that with probability 0.95, a new observation from the same group will fall inside the ellipse. Genotypes are indicated at the right of the figure. N = 256 data points. In (B) Heatmaps of codon usage in Spike proteins are shown. Unit variance scaling was applied. Each column corresponds to a different Spike protein from SARS-CoV-2 VOCs strains, who's genotype is shown in the upper part of the figure. Both rows and columns are clustered using correlation distance and average linkage.