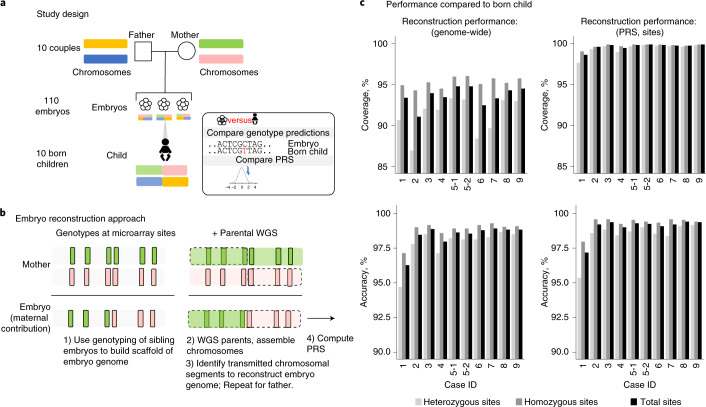
Fig. 1. WGR and approach.
a, This research study involved reconstruction of 110 embryo genomes from 10 couples and comparison to the genome sequence of the born child. Twelve PRS models were computed from the born-child samples and the 10 corresponding reconstructed embryos and compared for concordance. b, WGR involves whole-genome sequencing (WGS) of prospective parents and single-nucleotide polymorphism (SNP) microarray genotyping of sibling embryos (Methods and Supplemental Note 1). Allele measurements at each SNP are color-coded based on the parental haplotype of origin illustrated in a. A combination of molecular and statistical/population-based techniques phase the parents’ chromosomes, infer the locations of meiotic recombination for each embryo and correct errors introduced in the process of testing single-cell or few-cell embryo biopsies (Methods). Reconstructed embryo whole genomes are used to predict common disease risk by calculating PRSs and inferring the inheritance of rare variants with high impact on disease risk. c, Performance by comparing genotypes from WGR with the born child’s DNA shows genotype accuracies ranging from 99.0% to 99.4% at sites used in polygenic prediction in day-5 embryos and 97.2% to 99.1% in day-3 embryos. Case 1 includes only day-3 embryos, and case 2 includes both day-3 and day-5 embryos. All other cases included day-5 embryos only. Statistics are subdivided by genotype (heterozygous or homozygous) in the born child.