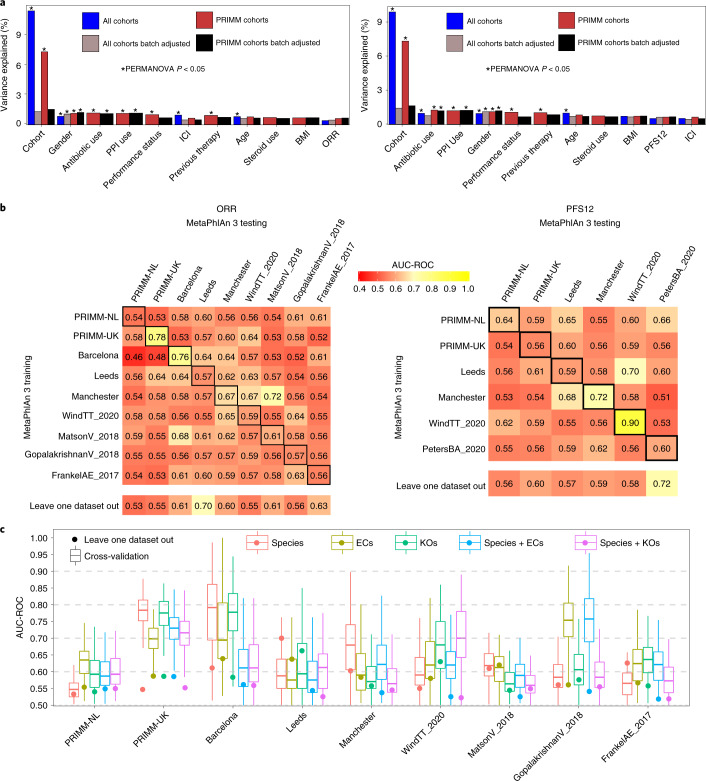
Fig. 2. Integrated analysis of newly sequenced and publicly available datasets for cross-cohort response–microbiome association.
a, Contribution of variables to the overall microbial community composition highlights the heterogeneity of the microbiome structure across cohorts that has a substantially higher effect than both anthropometric and clinical parameters. We either used all available cohorts or newly sequenced cohorts for which additional metadata were available. Batch-correction methods were applied to species-level abundances prior to distance calculations. The plot on the left uses ORR as the outcome variable, whereas the plot on the right adopts PFS12. b, Prediction matrix for microbiome-based prediction of response assessed via ORR (left matrix) and PFS12 (right matrix) within each cohort (values on the diagonal), across pairs of cohorts (one cohort used to train the model and the other for testing) and in the leave-one-cohort-out setting (training the model on all but one cohort and testing on the left-out cohort). We report the AUC-ROC values obtained from Lasso models on species-level relative abundances. Values on the diagonal refer to the median AUC-ROC values of 100-repeated fivefold-stratified cross-validations. Off-diagonal values refer to AUC-ROC values obtained by training the classifier on the cohort of the corresponding row and applying it to the cohort of the corresponding column. The leave-one-out row refers to the performances obtained by training the model using all but the cohort of the corresponding column and applying it to the cohort of the corresponding column. The same prediction matrix using functional microbiome profiles are available in Extended Data Fig. 4. c, ORR (n = 284) cross-validation AUC-ROC values obtained from Lasso models trained using 100-repeated fivefold-stratified cross-validations (boxplots) and leave-one-dataset-out AUC-ROC values from Lasso models obtained by training the model using species-level relative abundances and all but the corresponding (circles). The lower and upper hinges of boxplots correspond to the 25th and 75th percentiles, respectively. The midline is the median. The upper and lower whiskers extend from the hinges to the largest (or smallest) value no further than 1.5× interquartile range from the hinge, defined as the distance between the 25th and 75th percentiles. EC, enzyme category.