Abstract
Nucleotide second messengers, such as cAMP and c-di-GMP, regulate many physiological processes in bacteria, including biofilm formation. There is evidence of cross-talk between pathways mediated by c-di-GMP and those mediated by the cAMP receptor protein (CRP), but the mechanisms are often unclear. Here, we show that cAMP-CRP modulates biofilm maintenance in Shewanella putrefaciens not only via its known effects on gene transcription, but also through direct interaction with a putative c-di-GMP effector on the inner membrane, BpfD. Binding of cAMP-CRP to BpfD enhances the known interaction of BpfD with protease BpfG, which prevents proteolytic processing and release of a cell surface-associated adhesin, BpfA, thus contributing to biofilm maintenance. Our results provide evidence of cross-talk between cAMP and c-di-GMP pathways through direct interaction of their effectors, and indicate that cAMP-CRP can play regulatory roles at the post-translational level.
Subject terms: Bacterial genetics, Biofilms, Bacterial adhesion
Nucleotide second messengers, such as cAMP and c-di-GMP, regulate many physiological processes in bacteria, including biofilm formation. Here, the authors provide evidence of cross-talk between cAMP and c-di-GMP pathways through direct interaction of their effectors, showing that the cAMP receptor protein (CRP) can play regulatory roles at the post-translational level.
Introduction
The cAMP receptor protein (CRP) is a global transcription factor known to control the transcription of numerous genes by responding to changes in the intracellular cAMP level in most bacteria1. Extensive genetic, biochemical, biophysical, and structural data have been analyzed to determine how the cAMP-CRP complex controls gene transcription1–4. In Escherichia coli, apo-CRP (homodimer CRP in the absence of cAMP) is in the “off” state, which binds DNA nonspecifically and weakly5,6. On binding cAMP in the N-terminal domain, CRP undergoes an allosteric transition and is activated to the “on” state, which binds DNA specifically and strongly via its C-terminal domain6. CRP can directly interact with RNA polymerase or other transcription factors to control the transcription of corresponding genes, and both interactions are promoter-dependent7–9. Although known for its role during carbon catabolite repression, the biological role of cAMP-CRP goes far beyond carbon catabolite repression in most bacteria and includes toxin production10, iron acquisition11, capsule production12, and quorum sensing13. Thus, the transcription of numerous genes is controlled by cAMP-CRP in bacteria. Indeed, in E. coli, the transcription of more than 7% genes is regulated by cAMP-CRP14,15. Although most subsequent studies focused on cAMP-CRP as a transcription factor, cAMP-CRP may play a non-transcriptional regulatory role in some bacteria. For instance, in Mycobacterium tuberculosis, CRP may act as a nucleoid associated protein (NAP) to influence the dynamic spatial arrangement of the chromosome in a cAMP-independent manner16–18. However, in γ-proteobacteria, the researches on cAMP-CRP as a non-transcriptional regulator involved in physiological metabolism are limited.
Biofilms are structured communities of sessile, microbial cells encased in a self-secreted extracellular matrix, which is composed of exopolysaccharides, proteinaceous adhesin factors and nucleic acids19–21. The bacterial biofilm developmental process includes four stages: (i) initial attachment, (ii) microcolony formation, (iii) biofilm maturation, and (iv) dispersion22,23. There has been an abundance of studies on the regulation of initial attachment in multiple bacteria, less is known about dispersion. To investigate the transition from biofilm maturation to dispersion, the term “biofilm maintenance” has been proposed to describe the process by which existing mature biofilms regulate themselves to persist on a surface before dispersion24. The intracellular second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is involved in regulating each stage of biofilm development24–27. Diguanylate cyclase (DGC), containing a conserved GGDEF domain, catalyzes two molecules of GTP to synthesize c-di-GMP, which is degraded to 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) and/or GMP by phosphodiesterase (PDE) with a conserved EAL/HD-GYP domain28–30. Most bacteria have more than one DGC/PDE, the quantity of which is associated with the complexity of the habitat of bacteria31. For example, free-living microbes tend to have more DGC/PDE than obligate pathogenic bacteria31–33. Although most bacteria have multiple DGCs/PDEs, only a few DGCs/PDEs influence biofilm development at a defined time period29,32,34–37. Under most conditions, c-di-GMP is involved in regulating specific regulatory networks by forming a complex with c-di-GMP receptors/effectors38–41, which switch bacterial transition between sessile biofilm and planktonic modes depending on their ability to bind c-di-GMP25,37,39. As a consequence, a high intracellular c-di-GMP level is associated with biofilm formation, while a low intracellular c-di-GMP level tends to facilitate a planktonic lifestyle31,42,43.
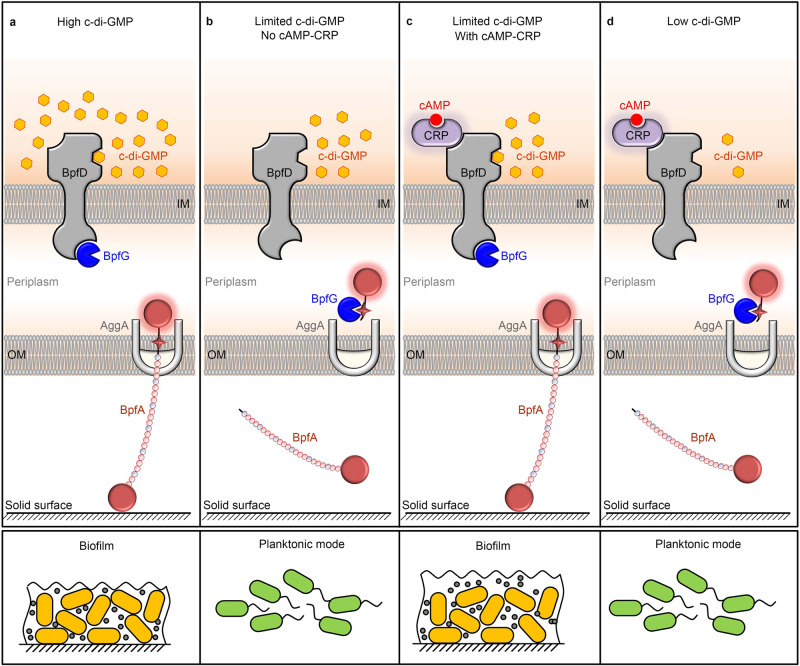
Shewanella species are gram-negative facultative anaerobic γ-proteobacteria, which are dissimilatory metal-reducing bacteria and widely distributed in aquatic niches44,45. The respiratory diversity and ability to form biofilms of Shewanella species allow for its use in various bioremediations and biotechnologies46–48. Previous studies have reported that the outer membrane adhesin BpfA promotes the biofilm formation of Shewanella oneidensis MR-1 and Shewanella putrefaciens CN32 through improving the cell adhesion to a solid surface49–52. Moreover, bpfA (Sputcn32_3591) is the first gene in an operon (Supplementary Fig. 1a) containing seven genes encoding a type I secretion system for the translocation of BpfA protein (Sputcn32_3592, Sputcn32_3593, AggA, Sputcn32_3595), a periplasmic protease BpfG (Sputcn32_3596), and an inner membrane-spanning c-di-GMP effector BpfD (Sputcn32_3597)49,53,54. Both BpfG and BpfD control whether BpfA is localized on the cell surface, and together, is defined as the BpfAGD system55. The regulation model of the BpfAGD system is similar to the Lap system of Pseudomonas fluorescens Pf0-142. As shown in Fig. 1, a high intracellular c-di-GMP level activates the c-di-GMP effector BpfD to bind to and sequester the periplasmic protease BpfG, which prevents BpfA being processed and results in biofilm formation (Fig. 1a). When intracellular c-di-GMP level is low, BpfD cannot sequester BpfG, leaving BpfG free to process and release BpfA from the cell surface, leading to planktonic mode55.
Fig. 1. Pattern for BpfAGD system regulated by cAMP and c-di-GMP.
a A high intracellular c-di-GMP level activates the c-di-GMP effector BpfD to bind to and sequester the periplasmic protease BpfG, which prevents BpfA being processed and results in biofilm formation. b When intracellular c-di-GMP level is limited, BpfD cannot sequester BpfG, leaving BpfG free to process and release BpfA from the cell surface, leading to planktonic mode. c Although intracellular c-di-GMP level is limited, cAMP and c-di-GMP synergistically maintain the interaction between BpfD and BpfG through the direct interaction of their effector proteins, CRP and BpfD, thereby supporting biofilm maintenance. d When the intracellular c-di-GMP content is low and below a certain threshold concentration, the presence of cAMP-CRP cannot maintain the interaction between BpfD and BpfG, leading to biofilm dispersion. IM: Inner membrane, OM: Outer membrane.
cAMP-CRP has been reported to regulate biofilm formation in some bacteria. For example, in Pseudomonas aeruginosa, a solid surface signal activates two adenylate cyclases, CyaA and CyaB, thereby promoting cAMP synthesis56. Subsequently, cAMP activates the transcriptional regulatory activity of Vfr, a homologous protein of CRP, and the Vfr-cAMP complex triggers the transcription of a range of genes that are involved in biofilm formation57,58. In addition, as a global transcription factor, cAMP-CRP has been reported to regulate the transcription of some genes encoding c-di-GMP receptors/effectors or DGCs/PDEs to control biofilm formation in some bacteria59–61. Compared to c-di-GMP, which acts as a “switch molecule” to control the transition between motile planktonic and sessile biofilm lifestyles in many bacteria, the function of cAMP-CRP in biofilm formation seems more ancillary62. Thus, the available research on the underlying mechanisms of cAMP-CRP regulating biofilm formation is limited.
In this work, we show that cAMP-CRP is required to maintain a mature biofilm of S. putrefaciens CN32. Further investigations indicate that cAMP-CRP physically interacts with the inner membrane-spanning c-di-GMP effector BpfD, and this interaction greatly enhances the capability of BpfD to interact with and sequester BpfG, thereby retaining BpfA on the cell surface and supporting biofilm maintenance. This report not only reveals that cAMP and c-di-GMP synergistically regulate biofilm maintenance through the direct interaction of their effectors but also describes a regulatory pattern that cAMP-CRP modulates biofilm maintenance acting as a post-translation regulator.
Results
cAMP-CRP complex regulates biofilm maintenance
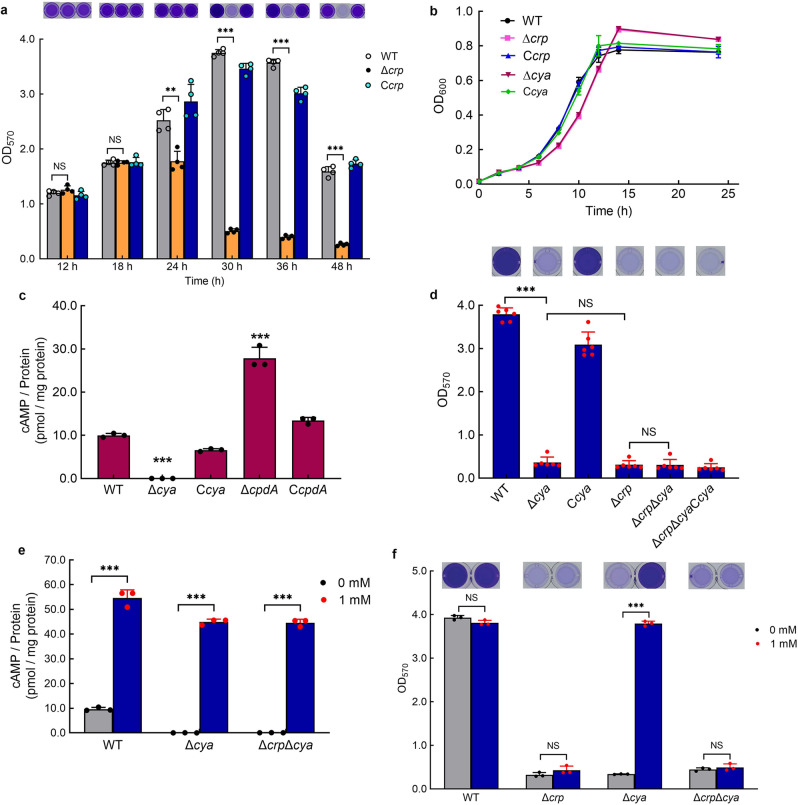
Genes associated with biofilm maintenance were screened using a transposon insertion mutation technique and crp (Sputcn32_0652) was identified. The biofilm assay showed that the biofilm biomasses of WT, Δcrp, and Ccrp were similar at 12 h; however, the Δcrp biofilm dispersed at 30 h, while the WT and Ccrp maintained a robust biofilm (Fig. 2a). The growth rates of the three strains showed that compared to WT and Ccrp, the growth rate of Δcrp was slightly slower at the early exponential phase (before 12 h), but Δcrp grew to a slightly higher cell density than the other strains after 12 h (Fig. 2b). The ratio of biofilm biomass to cell growth (OD570/OD600) of Δcrp was still significantly lower than that of the WT and Ccrp at 30 h (Supplementary Fig. 1b), suggesting that the statistically significant decline of the biofilm biomass of Δcrp was not caused by the change in cell growth. These results indicate that CRP is necessary to support the biofilm maintenance of S. putrefaciens CN32.
Fig. 2. cAMP-CRP complex supports biofilm maintenance in S. putrefaciens CN32.
a Biofilm biomass (n = 4 independent samples). b Cell growth (n = 3 independent samples). c Intracellular cAMP concentration at 30 h (n = 3 independent samples). d Biofilm biomass at 30 h (n = 6 independent samples). e Intracellular cAMP concentration with the addition of 1 mM exogenous cAMP to the culture medium vs. control (no addition of exogenous cAMP [0 mM]) at 30 h (n = 3 independent samples). f Biofilm biomass with the addition of 1 mM exogenous cAMP to the culture medium vs. control at 30 h (n = 3 independent samples). Insets in (a, d, f) are the biofilm pictures of crystal violet dyeing. Data in (a–f) are shown as the mean ± SD. Two-sided Student’s t test was used in (a, c–f) to analyze the statistical significance (NS: No significance. ***p < 0.001). Source data are provided as a Source Data file.
CRP is ubiquitous in bacteria and is widely known as a cAMP-dependent transcription factor. S. putrefaciens CN32 has three adenylate cyclases: CyaA (Sputcn32_3586), CyaB (Sputcn32_3104), and CyaC (Sputcn32_1140). In order to determine whether CRP regulates biofilm maintenance through forming a complex with cAMP, we created a mutant lacking all three adenylate cyclases ΔcyaAΔcyaBΔcyaC (named Δcya) and its complementation strain bearing a plasmid encoding all three adenylate cyclases CcyaA-CcyaB-CcyaC (named Ccya). The intracellular cAMP concentration was not detected in Δcya, and the intracellular cAMP concentration in Ccya was restored (Fig. 2c), which confirmed that Δcya is a cAMP-negative mutant. The biofilm assay showed that similar to Δcrp, the Δcya exhibited poor biofilm maintenance (Fig. 2d), while the deletion and complementation of three adenylate cyclase genes in Δcrp (ΔcrpΔcya and ΔcrpΔcyaCcya) did not influence the biofilm phenotype of Δcrp at 30 h (Fig. 2d). Meanwhile, the biofilm biomasses of WT, Δcya, Ccya, Δcrp, ΔcrpΔcya, and ΔcrpΔcyaCcya were similar at 12 h (Supplementary Fig. 1c). These results indicate that cAMP-CRP acts as a complex to regulate biofilm maintenance. The cell growth of Δcya and Δcrp were similar (Fig. 2b), indicating that the biofilm differences between WT and Δcya were not caused by the change in cell growth. Thus, the biofilm maintenance of S. putrefaciens CN32 at 30 h was supported by the cAMP-CRP complex.
We deleted the cAMP PDE gene cpdA (ΔcpdA) to further confirm that CRP regulates biofilm maintenance dependent on cAMP. Although the deletion of cpdA increased the intracellular cAMP concentration (Fig. 2c), the significant decrease in the cell growth of ΔcpdA (Supplementary Fig. 1d) made it difficult to analyze the biofilm phenotype. Thus, to further investigate the influence of the increasing intracellular cAMP concentration on biofilm maintenance, 1 mM exogenous cAMP was added to the biofilm medium. The results showed that the addition of exogenous cAMP increased the intracellular cAMP concentration of all tested strains (Fig. 2e); however, the increase in the intracellular cAMP concentration only restored the biofilm maintenance of Δcya, and exerted no effect on biofilm maintenance of Δcrp and ΔcrpΔcya at 30 h (Fig. 2f). These results further confirm that the biofilm maintenance is regulated by the cAMP-CRP complex.
cAMP-CRP maintains biofilm independently of its regulation of bpfA transcription
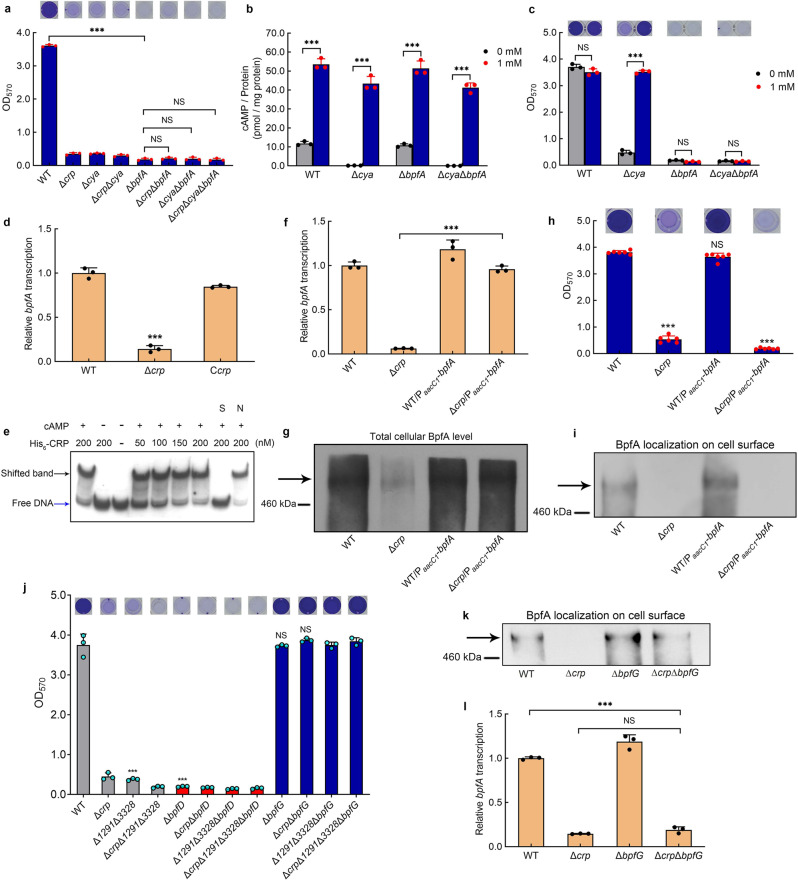
We next focused on investigating how cAMP-CRP regulates biofilm maintenance. It has been reported that large adhesive proteins play a critical role in biofilm formation or dispersion in several bacteria, including LapA in P. fluorescens and BpfA homolog in Shewanella spp.25,42. Previous studies have shown that S. putrefaciens CN32 exhibits defective biofilm formation due to the deletion of bpfA51,52. In this report, as Δcya, Δcrp and ΔcrpΔcya exhibit similar biofilm biomasses to ΔbpfA at 30 h (Fig. 3a), we considered that whether bpfA is the target regulated by cAMP-CRP. Therefore, we deleted bpfA in these three mutants, and the result showed that the biofilm biomasses of ΔcyaΔbpfA, ΔcrpΔbpfA and ΔcrpΔcyaΔbpfA were similar to that of ΔbpfA (Fig. 3a). Furthermore, we found that although the addition of exogenous cAMP increased the intracellular cAMP concentration in all of the tested strains (Fig. 3b), the increase in intracellular cAMP concentration only restored the biofilm biomass of Δcya to the WT level, but exerted no effects on the biofilm biomass of ΔcyaΔbpfA (Fig. 3c), indicating that the biofilm formation is poor as a result of the deletion of bpfA, irrespective of the change in intracellular cAMP concentration. These results suggest that BpfA is the downstream target of cAMP-CRP.
Fig. 3. cAMP-CRP maintains biofilm independently of its regulation of bpfA transcription.
a Biofilm biomass (n = 3 independent samples). b Intracellular cAMP concentration with adding 1 mM exogenous cAMP to the culture medium vs. control (n = 3 independent samples). c Biofilm biomass with adding 1 mM exogenous cAMP to the culture medium vs. control (n = 3 independent samples). d Transcriptional analysis of bpfA (n = 3 independent samples). e EMSA of cAMP-CRP binding to bpfA promoter (bpfA-pro). The His6-CRP (Lane–), labeled probe was incubated in the absence of His6-CRP. The concentrations of His6-CRP are shown above the figure. The binding specificity was confirmed by competitive assays with a 300-fold excess of unlabeled specific probe bpfA-pro (lane S) or unlabeled nonspecific competitor DNA (probe recA) (lane N). The cAMP (Lane –), labeled probe was incubated in the absence of cAMP. The cAMP (Lane + ), labeled probe was incubated in 1 μM cAMP. f The comparison of bpfA transcription between the native bpfA promoter and the constitutive promoter PaacC1 (n = 3 independent samples). g Western blotting detection of the total content of cellular BpfA protein. h Biofilm biomass (n = 6 independent samples). i Western blotting detection of BpfA localization on the cell surface. j Biofilm biomass (n = 3 independent samples). k Western blotting detection of BpfA localization on the cell surface. l Transcriptional analysis of bpfA (n = 3 independent samples). All strains used in (a–d, f–l) were cultured for 30 h in the biofilm state. Insets in (a, c, h, j) are the biofilm pictures of crystal violet dyeing. Data in (a–d, f, h, j, l) are shown as the mean ± SD. Two-sided Student’s t test was used in (a–d, f, h, j, l) to analyze the statistical significance (NS: No significance. ***p < 0.001). Source data are provided as a Source Data file.
In bacteria, cAMP-CRP is known as a global transcription factor. Thus, we considered whether cAMP-CRP directly controls the transcription of bpfA. Compared to WT and Ccrp, the transcription of bpfA in Δcrp was significantly down-regulated (Fig. 3d). The results of the electrophoretic mobility shift assay (EMSA) showed that in the presence of cAMP, CRP binds to the promoter region of bpfA (Fig. 3e), indicating that cAMP-CRP directly triggers the transcription of bpfA. To further investigate whether cAMP-CRP supports biofilm maintenance through triggering the transcription of bpfA, the native promoter of the bpfA operon was replaced by a constitutive promoter PaacC1 in WT and Δcrp. The result showed that the transcription of bpfA in Δcrp/PaacC1-bpfA was similar to that of WT and WT/PaacC1-bpfA, and significantly higher than that in Δcrp (Fig. 3f), which is consistent with their total intracellular BpfA protein levels (Fig. 3g). If cAMP-CRP regulates biofilm maintenance dependent on promoting the transcription of bpfA, the biofilm biomass of Δcrp/PaacC1-bpfA should be higher than that of Δcrp and close to that of WT. However, in practice, we found that increased transcription of bpfA in Δcrp/PaacC1-bpfA did not restore biofilm maintenance. The biofilm biomass of Δcrp/PaacC1-bpfA was similar to that of Δcrp and was still significantly lower than that of WT and WT/PaacC1-bpfA at 30 h (Fig. 3h), which was consistent with the localization of BpfA on the cell surface (Fig. 3i, Supplementary Fig. 1e). Thus, cAMP-CRP regulates the biofilm maintenance independently of its regulation of bpfA transcription.
We next investigate whether cAMP-CRP controls biofilm maintenance by retaining BpfA on the cell surface. As the retention of BpfA depends on the BpfAGD system (Fig. 1a), we deleted bpfD and bpfG in both WT and Δcrp. Similar to ΔbpfA, neither ΔbpfD nor ΔcrpΔbpfD was able to form biofilm, while the biofilm biomasses of ΔbpfG and ΔcrpΔbpfG at 30 h were similar to that of WT, and significantly higher than that of Δcrp (Fig. 3j). This finding suggests that the loss of BpfG restores the biofilm maintenance of Δcrp by preventing cleavage of BpfA from the cell surface. This was further confirmed by the immunoblot result showing that the localization of BpfA on the cell surface in ΔcrpΔbpfG was restored to the WT level, which was significantly higher than that in Δcrp (Fig. 3k, Supplementary Fig. 1f). In addition, we found that the transcription of bpfA in ΔcrpΔbpfG is similar to that in Δcrp (Fig. 3l), demonstrating that the difference in BpfA localization on the cell surface of Δcrp and ΔcrpΔbpfG is not caused by the loss of BpfG affecting bpfA transcription. Taken together, these findings show that cAMP-CRP supports biofilm maintenance through retaining BpfA on the cell surface, rather than through regulating bpfA transcription.
cAMP-CRP maintains biofilm independently of its regulation of c-di-GMP level
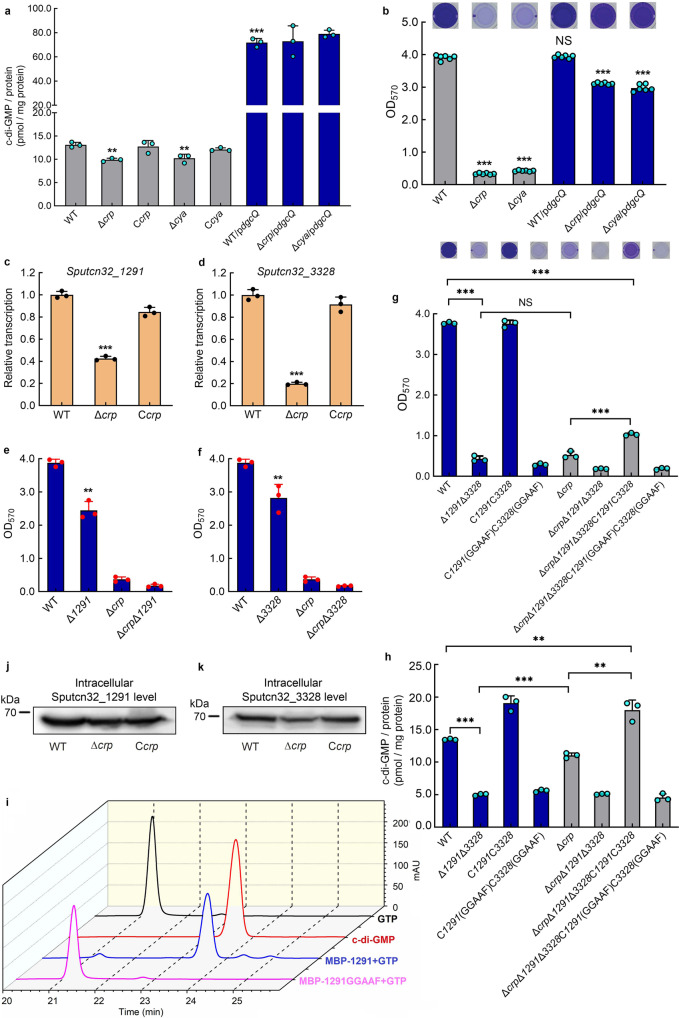
Previous studies have shown that the intracellular c-di-GMP level controls BpfA localization on the cell surface by regulating the BpfAGD system55 (Fig. 1a). Therefore, we considered whether cAMP-CRP controls biofilm maintenance through regulating the intracellular c-di-GMP level. Our results showed that compared to WT, the intracellular c-di-GMP concentration in Δcrp and Δcya decreased by ~25% (Fig. 4a). To test whether this 25% c-di-GMP decrease can cause biofilm dispersion of Δcrp and Δcya, we expressed DgcQ (formerly known as YedQ), a DGC of E. coli MG165563, in Δcrp and Δcya. The result showed that the introduction of DgcQ not only significantly increased intracellular c-di-GMP concentration in both strains (Fig. 4a) but also largely restored biofilm maintenance (Fig. 4b). In addition, intracellular cAMP was still not detected in Δcya/pdgcQ (Supplementary Fig. 1g), indicating that the biofilm maintenance restoration of Δcrp/pdgcQ and Δcya/pdgcQ is due to the increase in intracellular c-di-GMP concentration, rather than the intracellular cAMP concentration. These results suggest that cAMP-CRP seems to modulate biofilm maintenance by regulating the transcription levels of some dgc/pde genes.
Fig. 4. cAMP-CRP maintains biofilm independently of its regulation of c-di-GMP level.
a Intracellular c-di-GMP concentration (n = 3 independent samples). b Biofilm biomass (n = 6 independent samples). c, d Transcriptional analysis of Sputcn32_1291 and Sputcn32_3328 (n = 3 independent samples). e, f, g Biofilm biomass (n = 3 independent samples). h Intracellular c-di-GMP concentration (n = 3 independent samples). All strains used in (a–h) were cultured for 30 h in the biofilm state. i DGC enzymatic assays showing the DGC activity of Sputcn32_1291, GTP and c-di-GMP were used as standard samples in HPLC. j, k Western blotting detection of the total protein content of cellular Sputcn32_1291 and Sputcn32_3328 at 30 h. Insets in b, g are the biofilm pictures of crystal violet dyeing. Data in (a–h) are presented as the mean ± SD. Two-sided Student’s t test was used in (a–h) to analyze the statistical significance (NS: No significance. **p < 0.01. ***p < 0.001). Source data are provided as a Source Data file.
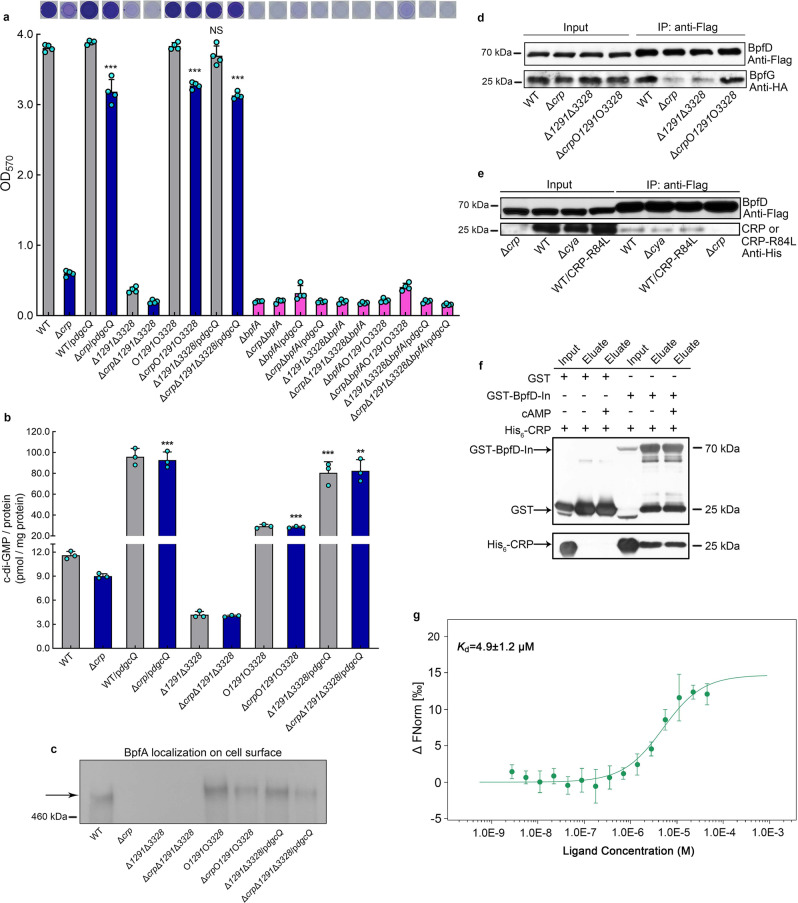
To screen biofilm maintenance-related dgc/pde genes whose transcriptions are regulated by cAMP-CRP, the transcription levels of all 47 dgc/pde genes in the genome of S. putrefaciens CN32 were compared in WT and Δcrp at 30 h (Fig. 4c, d, Supplementary Table 1). The results showed that cAMP-CRP regulates the transcription of 41 of the dgc/pde genes. To increase screening efficiency, we selected 11 genes whose transcription level in Δcrp changed by more than 2.5-fold compared to WT, and only the biofilm biomasses of Δ1291 and Δ3328 were slightly lower than that of the WT at 30 h (Fig. 4e, f, Supplementary Fig. 2). The biofilm of the double mutant Δ1291Δ3328 was poor, while the WT and C1291C3328 maintained robust biofilms at 30 h (Fig. 4g). And the intracellular c-di-GMP concentration in Δ1291Δ3328 was significantly lower than that in the WT and C1291C3328 (Fig. 4h). Moreover, the intracellular c-di-GMP concentration and the biofilm biomass of a site-directed mutant C1291(GGAAF)C3328(GGAAF) were similar to those of Δ1291Δ3328 at 30 h (Fig. 4g, h). These results indicate that Sputcn32_1291 and Sputcn32_3328 have DGC activity in vivo and participate in supporting biofilm maintenance. Besides, the results of biochemical assays showed that Sputcn32_1291 could catalyze GTP to produce c-di-GMP in vitro (Fig. 4i). However, we failed to detect the DGC or PDE activity of Sputcn32_3328 in vitro, which may be due to Sputcn32_3328 belongs to a three-component regulatory system, DGC activity of Sputcn32_3328 may depend on its cognate proteins in vivo. We confirmed that both Sputcn32_1291 and Sputcn32_3328 act as DGCs in vivo to regulate biofilm maintenance; this is not only due to the presence of the conserved GGDEF domain of DGC activity in both proteins, which influence the intracellular c-di-GMP concentration, but also due to the fact that the heterologous expression of dgcQ in Δ1291Δ3328 restored the biofilm biomass to the WT level (Fig. 5a) by increasing the intracellular c-di-GMP concentration (Fig. 5b).
Fig. 5. Direct interaction between cAMP-CRP and BpfD retains BpfA localization on the cell surface.
a Biofilm biomass (n = 3 independent samples). b Intracellular c-di-GMP concentration (n = 3 independent samples). c Western blotting detection of BpfA localization on the cell surface. d Co-IP to analyze the interaction between BpfD and BpfG in vivo. e Co-IP to analyze the interaction between BpfD and CRP/CRP-R84L in vivo. All strains used in (a–e) were cultured for 30 h in the biofilm state. f GST pull-down assay showing the interaction between BpfD intracellular domains and CRP in vitro. g MST showing the interaction between BpfD intracellular domains and CRP in vitro, data are shown as the mean ± SD (n = 3 independent samples). The native promoter region of the bpfA operon of the strains used in (c–e) was replaced by the constitutive promoter PaacC1. Insets in (a) are the biofilm pictures of crystal violet dyeing. Data in (a, b) are shown as the mean ± SD. Two-sided Student’s t test was used in (a, b) to analyze the statistical significance (NS: No significance. **p < 0.01. ***p < 0.001). Source data are provided as a Source Data file.
As cAMP-CRP regulates the transcription of Sputcn32_3328 and Sputcn32_1291 genes (Fig. 4c, d) and Sputcn32_1291 and Sputcn32_3328 act as DGCs to support biofilm maintenance (Fig. 4g, h), it seems that cAMP-CRP regulates biofilm maintenance through modulating the intracellular c-di-GMP level. However, Fig. 4g, h show some contradictory results. First, the intracellular c-di-GMP concentration of Δcrp was significantly higher than that of Δ1291Δ3328 (Fig. 4h); however, both Δcrp and Δ1291Δ3328 showed similarly poor biofilm maintenance, which was significantly lower than that of the WT (Fig. 4g). Second, if cAMP-CRP regulated biofilm maintenance by modulating the intracellular c-di-GMP level, the increase in intracellular c-di-GMP concentration of Δcrp to the WT level should restore its biofilm biomass to the WT level. However, in practice, we found that the intracellular c-di-GMP concentration in ΔcrpΔ1291Δ3328C1291C3328 was significantly higher than that in Δcrp, and even higher than that of the WT (Fig. 4h), but its biofilm biomass was slightly higher than that of Δcrp, and still significantly lower than that of WT (Fig. 4g). Thus, the biofilm maintenance supported by cAMP-CRP may not be totally dependent on regulating the intracellular c-di-GMP level. Besides, although the transcriptional levels of Sputcn32_1291 and Sputcn32_3328 genes in Δcrp were significantly lower than those in WT at 30 h (Fig. 4c, d), the intracellular level of both proteins at 30 h showed no significant difference or only changed slightly between WT and Δcrp (Fig. 4j, k). Thus, cAMP-CRP regulates biofilm maintenance independently of its regulation of the transcription of both dgc genes. The finding that the expression of DgcQ in Δcrp can partly restore its biofilm maintenance (Fig. 4b) is probably due to DgcQ significantly enhancing the intracellular c-di-GMP concentration (Fig. 4a), which acts as a remedial mechanism in the absence of cAMP-CRP.
To further confirm this hypothesis, we significantly increased the intracellular c-di-GMP concentration of Δ1291Δ3328 and ΔcrpΔ1291Δ3328 through over-expressing dgcQ in both mutants (Fig. 5b). The biofilm assay showed that only the biofilm biomass of Δ1291Δ3328/pdgcQ was restored to the WT level, but the biofilm biomass of ΔcrpΔ1291Δ3328/pdgcQ was still lower than that of WT (Fig. 5a), indicating that the deletion of crp significantly weakened the restoration effect of increased the intracellular c-di-GMP level on biofilm maintenance. Taken together, although increasing the intracellular c-di-GMP level can partly restore the decrease in biofilm maintenance caused by the absence of cAMP-CRP, cAMP-CRP regulates biofilm maintenance independently of its regulation of the intracellular c-di-GMP concentration.
cAMP-CRP and c-di-GMP maintain biofilm by retaining BpfA on the cell surface
To further uncover the mechanism by which cAMP-CRP and c-di-GMP regulate biofilm maintenance, we deleted bpfA, bpfD and bpfG in both Δ1291Δ3328 and ΔcrpΔ1291Δ3328. The results showed that similar to the deletion of bpfA or bpfD in Δcrp, deletion of bpfA or bpfD blocked the biofilm formation of Δ1291Δ3328 and ΔcrpΔ1291Δ3328 (Figs. 3j, 5a), which is consistent with the regulatory pattern of the BpfAGD system as shown in Fig. 1a. Moreover, the deletion of bpfG restored the biofilm maintenance of Δcrp, Δ1291Δ3328 and ΔcrpΔ1291Δ3328 to the WT level (Fig. 3j), suggesting that both c-di-GMP and cAMP-CRP regulate biofilm maintenance through controlling the BpfAGD system.
To further confirm this finding, the influence of cAMP-CRP and c-di-GMP on the localization of BpfA on the cell surface was analyzed. We first replaced the native promoter region of the bpfA operon with the constitutive promoter PaacC1 in WT, Δcrp, Δ1291Δ3328, ΔcrpΔ1291Δ3328, O1291O3328, ΔcrpO1291O3328, Δ1291Δ3328/pdgcQ, and ΔcrpΔ1291Δ3328/pdgcQ to eliminate the differences in bpfA transcription caused by changes in the c-di-GMP and cAMP-CRP levels. The qRT-PCR results showed that the promoter replacement of the bpfA operon reduced the transcription differences in the bpfA operon among these eight strains (Supplementary Fig. 3a), but that the replacement did not change the biofilm maintenance phenotype of the corresponding strains (Supplementary Fig. 3b). We next examined the BpfA localization on the cell surface of strains with PaacC1-bpfA replacement using immunoblot. The result showed that BpfA was almost undetectable on the cell surface of Δcrp, Δ1291Δ3328, and ΔcrpΔ1291Δ3328 (Fig. 5c, Supplementary Fig. 3c), indicating that the absence of cAMP-CRP and the reduction in intracellular c-di-GMP concentration causes the release of BpfA from the cell surface. Besides, the increase in the intracellular c-di-GMP concentration retains the BpfA localization on the cell surface (see the mutants O1291O3328, ΔcrpO1291O3328, Δ1291Δ3328/pdgcQ and ΔcrpΔ1291Δ3328/pdgcQ in Fig. 5c, Supplementary Fig. 3c), thereby supporting biofilm maintenance (Fig. 5a). Thus, both cAMP-CRP and c-di-GMP support biofilm maintenance by retaining BpfA on the cell surface.
cAMP-CRP and BpfD interaction retains BpfA on the cell surface
Finally, we sought to investigate how cAMP-CRP retains BpfA on the cell surface. As shown in Fig. 1a, BpfA localization on the cell surface is regulated by the interaction between BpfD (c-di-GMP effector) and BpfG55. Thus, the interaction between BpfD and BpfG in WT, Δcrp, Δ1291Δ3328, and ΔcrpO1291O3328 was detected by co-immunoprecipitation (Co-IP). In all four strains, the native promoter region of the bpfA operon was replaced with the constitutive promoter PaacC1, a chromosomal C-terminal 3×Flag-tag was attached to BpfD, and a chromosomal near C-terminal 1×HA-tag was attached to BpfG, all of which did not influence the biofilm maintenance compared to that of their original strains (Supplementary Fig. 3d). After the replacement of bpfA operon promoter, the protein levels of BpfD and BpfG were similar in the four strains (input bands of Fig. 5d), indicating that the differences in BpfA localization on the cell surface among these strains were not caused by the change in total protein of BpfD and BpfG. The Co-IP result showed that the interaction between BpfD and BpfG was greatly weakened in Δcrp and Δ1291Δ3328 compared to that in WT and ΔcrpO1291O3328, indicating that both cAMP-CRP and c-di-GMP regulate the interaction between BpfD and BpfG (Fig. 5d, Supplementary Fig. 3e). This finding is consistent with the previously reported conclusion that c-di-GMP is essential in promoting the interaction between BpfD and BpfG55,64.
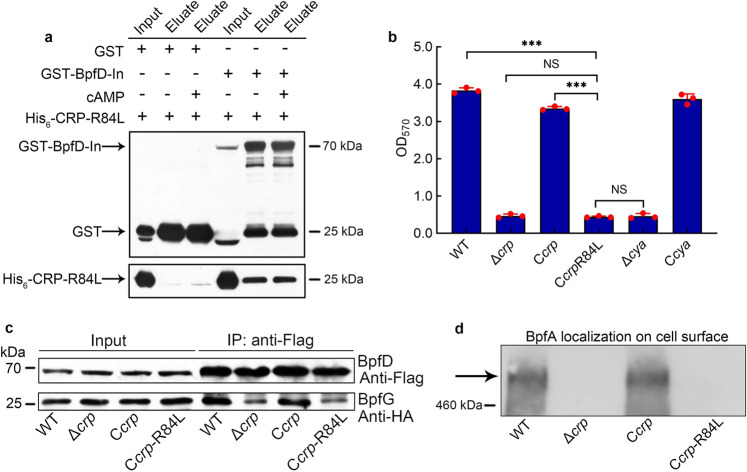
We next consider how cAMP-CRP regulates the interaction between BpfD and BpfG. It has been reported that GcbC, a DGC, enhances the interaction between LapD and LapG by directly binding to LapD in P. fluorescens65. This led us to consider that there may be a CRP-regulated protein or CRP-self, which acts as the enhancer to promote the interaction between BpfD and BpfG. The results of Co-IP, GST-pull down, and microscale thermophoresis (MST) experiments all showed that CRP directly interacted with BpfD, and that their interaction does not require the presence of cAMP (Fig. 5e–g, Supplementary Fig. 3f). However, as the above results indicate that cAMP-CRP acts as a complex to regulate biofilm maintenance (Fig. 2d, f), we considered whether cAMP is necessary to enhance the interaction between BpfD and BpfG. A site-directed mutant protein CRP-R84L (Arg-84→Leu), which has lost the ability to bind cAMP3 (Supplementary Fig. 3g), still interacted with BpfD (Figs. 5e, 6a). However, the interaction between BpfD and BpfG, BpfA localization on the cell surface, and the biofilm maintenance of Ccrp-R84L were similar to those in Δcrp (Fig. 6b–d, Supplementary Fig. 3h). These results indicate that although CRP physically interacts with BpfD (Fig. 5e), the ability of CRP to enhance the interaction between BpfD and BpfG is lost in the absence of cAMP (Fig. 6c, Supplementary Fig. 4a–c).
Fig. 6. cAMP is not necessary for physical interaction between CRP and BpfD, but it is essential to help CRP enhance the interaction between BpfD and BpfG.
a GST pull-down assay showing the interaction between BpfD intracellular domains and CRP-R84L in vitro. b Biofilm biomass (n = 3 independent samples). c Co-IP to analyze the interaction between BpfD and BpfG in vivo. d Western blotting detection of BpfA localization on the cell surface. All strains used in (a, c, d) were cultured for 30 h in the biofilm state. The native promoter region of the bpfA operon of the strains used in (c, d) was replaced by the constitutive promoter PaacC1. Data in (a) are shown as the mean ± SD. Two-sided Student’s t test was used in (a) to analyze the statistical significance (NS: No significance. ***p < 0.001). Source data are provided as a Source Data file.
Pattern for cAMP and c-di-GMP synergistically maintaining biofilm
Based on the above results, we propose a pattern to explain the phenotype of the different mutants. As shown in Fig. 1c, cAMP and c-di-GMP synergistically maintain the interaction between BpfD and BpfG through the direct interaction of their effector proteins, CRP and BpfD, thereby supporting biofilm maintenance. The presence (such as WT) or absence (such as Δcrp) of the cAMP-CRP complex determines the biofilm maintenance (Fig. 1c) or biofilm dispersion (Fig. 1b), respectively. Significant increase in the intracellular c-di-GMP concentration in Δcrp (such as ΔcrpO1291O3328, Fig. 1a) maintains the interaction between BpfD and BpfG (Fig. 5d), which retains BpfA on the cell surface (Fig. 5c, Supplementary Fig. 3c), thereby greatly remedying the biofilm maintenance of Δcrp (Fig. 5a). When the intracellular c-di-GMP content is low and below a certain threshold concentration (such as Δ1291Δ3328, Fig. 1d), the interaction between BpfD and BpfG cannot be maintained (Fig. 5d). As a result, BpfA is released from the cell surface (Fig. 5c, Supplementary Fig. 3c), leading to biofilm dispersion (Figs. 1d, 5a). Moreover, significant increase in the intracellular c-di-GMP concentration (Fig. 4a) exerts no effect on biofilm maintenance of WT (Fig. 4b); this is probably due to the fact that, in WT, as BpfG is already inhibited enough by BpfD in the presence of cAMP-CRP, further increasing the intracellular c-di-GMP level has no additive effect. The above conclusions are further confirmed by Supplementary Figs. 1g, 4d–f.
The relationships between intracellular c-di-GMP level and bacterial lifestyles have been well-established. Specially, a high intracellular c-di-GMP level is associated with biofilm formation, while a low intracellular c-di-GMP level tends to facilitate a planktonic lifestyle. However, several reports demonstrate that some regulators act as additional tool for fine-tuning such an important cellular molecular mechanism by cross-talking with c-di-GMP66. Our results reveal that in limited intracellular c-di-GMP level condition, cAMP-CRP plays a determinant function in the regulation of biofilm maintenance in S. putrefaciens CN32 (Fig. 1b, c), which underlines the complexity of bacterial second messenger regulation again. In summary, in S. putrefaciens CN32, cAMP-CRP acts as not only global transcription factor to regulate physiological metabolism but also post-translation regulator to participate in biofilm maintenance by cross-regulating with second messenger c-di-GMP.
Discussion
Bacteria have two growth modes: biofilm, which is considered as a dominant bacterial growth mode in nature, and planktonic growth26,67. In the biofilm developmental process, initial attachment and dispersion are widely referred to as promising avenues for biofilm control because both are key steps in the transition between motile planktonic and sessile biofilm lifestyles22,23. Although there has been an abundance of studies on the regulation of initial attachment in multiple bacteria, less is known about the maintenance of mature biofilm and dispersion29,67. Recently, two PDEs, RmcA and MorA, have been shown to regulate biofilm maintenance of P. aeruginosa, indicating that c-di-GMP plays a significant role in regulating biofilm maintenance24. Compared to c-di-GMP, the function of cAMP in biofilm formation seems more ancillary in most bacteria62. However, in this report, cAMP and c-di-GMP were found to play an equally important role and synergistically regulate biofilm maintenance in S. putrefaciens CN32, suggesting that the importance of cAMP in regulating biofilm development in bacteria may be underestimated in previous studies.
Several bacteria possess large adhesins localized on their cell surface, which assist with adherence to biotic or abiotic surfaces. Such adhesins are commonly controlled by a Lap system whose underlying mechanism has been well-established in P. fluorescens42. In S. putrefaciens CN32, a BpfAGD system that belongs to the Lap system controls biofilm formation by responding to intracellular c-di-GMP levels55. As shown in Fig. 1a, a high intracellular c-di-GMP level increases the interaction between BpfD and BpfG, which retains BpfA on the cell surface, thereby promoting initial biofilm formation51,52,55. Thus, we originally speculated that the cAMP-CRP complex may control biofilm maintenance by regulating the transcription of the bpfA operon and/or dgc/pde genes given that cAMP-CRP is widely recognized as a transcription factor. Indeed, the transcription of the bpfA operon and two key dgc genes in Δcrp were significantly lower than those in WT. However, further investigations showed that the reduction in the transcription of bpfA operon and dgc genes does not play a decisive role in the poor biofilm maintenance of Δcrp.
Further study showed that cAMP-CRP and c-di-GMP regulate BpfA localization on the cell surface. We next sought to determine how cAMP-CRP supports biofilm maintenance through regulating BpfAGD system. In P. fluorescens, the Lap system not only responds to the total intracellular c-di-GMP level, but is also specifically controlled by local c-di-GMP signaling42. GcbC is a DGC localized on the inner membrane, which contributes to the local c-di-GMP pool and is not responsible for the total intracellular c-di-GMP level. Citrate increases the c-di-GMP synthesis capability of GcbC and stimulates the physical interaction between GcbC and LapD65. Subsequently, GcbC synthesizes c-di-GMP and physically delivers c-di-GMP to LapD by contacting with LapD42,65. Thus, the Lap system responds to citrate signals and promotes biofilm formation in a GcbC-dependent manner42. The enhancement of the interaction between LapD and LapG by GcbC suggests that in S. putrefaciens CN32, there may be a CRP-regulated protein or CRP-self, which greatly enhances the interaction between BpfD and BpfG by directly contacting with BpfD. Surprisingly, through Co-IP, GST-pull down, and MST experiments, we found that CRP directly interacts with the c-di-GMP effector BpfD (Fig. 5e–g) and enhances the interaction between BpfD and BpfG in the presence of cAMP (Fig. 6c), thereby supporting biofilm maintenance. We describe a regulatory pattern that cAMP-CRP acts as a post-translation regulator to modulate biofilm maintenance.
When cAMP-CRP acts as a transcription factor, the amino acid residue of CRP binding to cAMP is Arg-83 in E. coli3, which is Arg-84 in S. putrefaciens CN32 (Supplementary Fig. 3g). In this report, we found that although the CRP-R84L protein still interacted with BpfD (Figs. 5e, 6a), its function in regulating biofilm maintenance was lost (Fig. 6b–d). This not only indicates that CRP and cAMP must form a complex to perform regulatory function but also suggests that cAMP-CRP complex functions in the same conformation irrespective of acting as a transcription factor or a post-translation regulator. Extensive experiments are required to prove this hypothesis in future studies. Besides, heterologous complementation CRP (88% sequence identity with CN32 CRP) of E. coli MG1655 in Δcrp can partly restore the biofilm maintenance, but it cannot restore the biofilm biomass in ΔcrpΔcya (Supplementary Fig. 4g), indicating that the homologous CRP in E. coli has a function similar to CN32 CRP. Taken together, the above findings may suggest that the regulatory pattern of CRP as post-translation regulator is ubiquitous in bacteria. Future studies should pay more attention to the ability of cAMP-CRP to act as a post-translation regulator in bacteria.
As the biofilm biomasses of WT and Δcrp (Δcya) were similar at 12 h (Fig. 2a, Supplementary Fig. 1c), it seems that cAMP-CRP did not participate in the early biofilm developmental stage. Similar to the Lap system of P. fluorescens, the BpfAGD system not only regulates initial attachment55 but also controls biofilm dispersion of S. putrefaciens CN32. If cAMP-CRP can participate in biofilm maintenance through regulating the BpfAGD system, why is cAMP-CRP not involved in the early biofilm developmental stage? In P. fluorescens Pf0-1, similar to GcbC, another DGC, GcbB, is also involved in controlling biofilm formation through interacting with LapD, indicating multiple proteins regulate biofilm formation depending on interaction with LapD42. Thus, we speculated that although the BpfAGD system is involved in the entire biofilm developmental process, its upstream regulator varies depending on the different biofilm developmental stages. CRP interacts with BpfD in the later stage, and there may be another GcbC-like protein that participates in the regulation of initial attachment. In future, extensive experiments are required to explore more upstream regulators of the BpfAGD system at different biofilm developmental stages.
In bacteria, multiple nucleotide second messengers, such as cAMP, c-di-GMP, and guanosine tetra- and pentaphosphate ((p)ppGpp), have been reported to participate in the modulation of fundamental physiological processes28. Moreover, various second messengers coordinate multiple physiological metabolisms by cross-regulating each other68,69. For instance, cAMP has been reported to regulate the transcription of some genes encoding c-di-GMP receptors or DGCs/PDEs59–61. Furthermore, c-di-GMP and (p)ppGpp competitively bind to a common effector to control Caulobacter crescentus transition between swarmer and stalked lifestyles70. In this study, we describe a regulatory pattern whereby cAMP and c-di-GMP synergistically regulate biofilm maintenance by interaction of their effectors, which enriches the cross-regulation patterns between multiple second messengers. These findings are of great significance for understanding how bacteria intersect and integrate signals of second messengers. In addition, cAMP is ubiquitous in animals, plants, and microbes, and is known as a universal second messenger of organisms. It has been reported that pathogenic bacteria have evolved strategies to manipulate host cAMP concentrations71. In turn, cAMP not only is synthesized by bacteria themselves but also can enter the bacterial cells from their live environments72. Therefore, this study will assist with understanding the interaction between bacteria biofilms and host organisms.
Methods
Bacterial strains and growth conditions
E. coli strains were grown in Luria-Bertani broth (LB) at 37 °C. S. putrefaciens CN32 WT and its derivative strains were cultured at 30 °C in LB broth or modified M1 defined minimal medium (MM1) containing 20 mM sodium lactate, 30 mM HEPES, 1.34 mM KCl, 28.04 mM NH4Cl, 4.35 mM NaH2PO4, 7.5 mM NaOH, and 0.68 mM CaCl2 supplemented with trace amounts of amino acids, minerals, and vitamins51. The cell growth of CN32 and its derivatives was tested in MM1 medium without addition 0.68 mM CaCl2. When necessary, 50 μg ml−1 kanamycin (Km) was supplemented in medium for CN32 derivatives. The strains and plasmids used in this study are listed in Supplementary Table 2, and the primers are listed in Supplementary Table 3. The kits used for the isolation and purification of DNA were purchased from Tiangen Biotech (China). The enzymes for molecular manipulation were purchased from New England BioLabs (USA) and Thermo Fisher Scientific (USA).
Transposon mutagenesis and mapping the transposon insertion
Transposon mutagenesis was performed by biparental conjugation between S. putrefaciens CN32 receptor and E. coli UQ3022 donor (harboring a plasmid pRL27)51,73. S. putrefaciens CN32 and E. coli UQ3022 were cultured in LB medium for 12 h. The cells were collected by centrifugation at 10,000 g for 30 s and washed twice using LB medium. Then, 50 µl mixtures of two strains were spot inoculated on LB agar plate. After incubation at 30 °C for 6 h, the cells were resuspended using 1 ml LB medium. A transposon insertion library was obtained by plating 100 μl of cell suspension on LB agar plate containing 20 μg ml−1 tellurite and 50 μg ml−1 kanamycin. Then, the plates were incubated at 30 °C for 36 h, and the black colonies were selected for biofilm formation assay. To identify the location of transposon insertion, the chromosomal DNA was extracted and digested with BamHI or SpeI. The resulting fragments were self-ligated and transformed into E. coli UQ3021 cells. Subsequently, the transposon junction plasmids were extracted from the selected transformants and sequenced using primers Tn5-seq1/Tn5-seq2 (Supplementary Table 3) to reveal the location of transposon insertion.
Construction of deletion mutants and complementation strains
In-frame deletion mutants were generated using an established method51. To construct the crp deletion mutant, a 1164-bp upstream fragment (–1032 to +132 bp relative to the crp start codon) and a 1174 bp downstream fragment (+625 to +1159 bp relative to the crp start codon) were amplified using (primer pairs crp-5F/crp-5R and crp-3F/crp-3R, respectively). The homologous arms were digested by EcoRI/XbaI and XbaI/HindIII, and both were cloned into the EcoRI/HindIII-digested vector pK19mobsacB74 to yield pK19crpUD. The pK19crpUD was introduced into S. putrefaciens CN32 by conjugation with the help of pRK201375. The Δcrp deletion mutant was verified by PCR, using the following primer pairs crp-UF/crp-DR, crp-OF/crp-DR, crp-UF/crp-OR, and crp-INF/crp-INR. A similar strategy was applied for generating Δcya, ΔcpdA, ΔbpfA, ΔbpfD, ΔbpfG, Δ0133, Δ0654, Δ1291, Δ1365, Δ1412, Δ1858, Δ1934, Δ3319, Δ3328, and Δ3598 deletion mutants. The complementation strains were generated using plasmid pBBR1MCS-251,76. All of the resulting mutants and complementation strains were verified by PCR and DNA sequencing.
Construction of tagged transformant and replacement of PbpfA
The nucleotide sequences encoding tags were inserted into the corresponding location of the target gene in the genome. To construct a C-terminal 3×Flag-tagged BpfD transformant, the 3′-terminus region of the bpfD gene, including its upstream and downstream flanks was amplified by the primers BpfD-C5F/BpfD-C3R and cloned into pK19mobsacB to yield an intermediate plasmid pK19-BpfD-Cter. The pK19-BpfD-Cter was linearized by PCR amplification using the primers BpfD-FlagKinF and BpfD-FlagKinR, and the yielding fragment was digested by XbaI and XhoI, defined as pK19-BpfD-Cter-XbaI-XhoI. A commercially synthesized nucleotide sequence GAT TAC AAG GAT GAC GAC GAT AAG GAC TAT AAG GAC GAT GAT GAC AAG GAC TAC AAA GAT GAT GAC GAT AAA encoding 3×Flag (DYKDDDDKDYKDDDDKDYKDDDDK) was used as a template and amplified by primers Flag-F/FlagbpfD-R. The yielding fragment was digested with XbaI and XhoI, which was cloned into pK19-BpfD-Cter-XbaI-XhoI to yield the BpfD C-terminal 3×Flag-tag knock-in plasmid, pK19-BpfD-Cter-FlagKin, which was conjugated into WT. Thus, the 3×Flag nucleotide sequence was inserted after bpfD in-frame by homologous recombination to yield WT/BpfD-Flag. The final transformant was verified by PCR, using the following primer pairs BpfD-CSF/BpfD-CSR, BpfD-COF/BpfD-CSR, and BpfD-CSF/BpfD-COR. A similar strategy was used to generate Δcrp/BpfD-Flag, Δcya/BpfD-Flag, Δ1291Δ3328/BpfD-Flag, and ΔcrpΔ1291Δ3328/BpfD-Flag. All transformants were verified by PCR and DNA sequencing. To construct a 3×Flag-tagged BpfA transformant, 3×Flag was inserted after residue 3700 aa (11100 bp) in the full-length protein of 4220 aa and the construction strategy was similar to that of WT/BpfD-Flag. The transformant was verified by PCR and DNA sequencing.
To construct a 1×HA-tagged BpfG transformant, HA was inserted after residue 221 aa (663 bp) in the full-length protein (235 aa)64. The construction strategy was as follows: the 3′-terminus region of bpfG, including its upstream and downstream flanks was amplified by the primers BpfG-C14-5F and BpfG-C14-3R and cloned into pK19mobsacB to yield an intermediate plasmid pK19-BpfG-Cter; pK19-BpfG-Cter was linearized by PCR amplification using the 5′-phosphorylated primers BpfG-HAKinF and BpfG-HAKinR; the linearized plasmid was self-ligated to yield the BpfG-HA knock-in plasmid, pK19-BpfG-C14-HAKin; and finally, the plasmid was conjugated into WT/BpfD-Flag to obtain the transformant WT/BpfD-Flag/BpfG-HA. The final transformants were verified by PCR, using the primers BpfG-C14-SF/BpfG-C14-SR, BpfD-C14-OF/BpfD-C14-SR, and BpfD-C14-SF/BpfD-C14-OR. A similar strategy was used to generate the transformants Δcrp/BpfD-Flag/BpfG-HA, Δcya/BpfD-Flag/BpfG-HA, Δ1291Δ3328/BpfD-Flag/BpfG-HA, and ΔcrpΔ1291Δ3328/BpfD-Flag/BpfG-HA. All transformants were verified by PCR and DNA sequencing.
To construct the 10×His-tagged CRP transformant, the 3′-terminus region of the crp gene, including its upstream and downstream flanks was amplified by primers CRP-C5F/CRP-C3R and cloned into pK19mobsacB to yield an intermediate plasmid pK19-CRP-Cter; pK19-CRP-Cter was linearized by PCR amplification using the primers CRP-His10KinF and CRP-His10KinR, both of which are 5′-phosphorylated primers containing the nucleotide sequence 10×CAT encoding 10×His. Next, the linearized plasmid was self-ligated to yield the CRP-His10 knock-in plasmid, pK19-CRP-C-His10Kin, which was conjugated into WT/BpfD-Flag/BpfG-HA to obtain the transformant WT/BpfD-Flag/BpfG-HA/CRP-His. The final transformants were verified by PCR, using the primer pairs CRP-CSF/CRP-CSR, CRP-COF/CRP-CSR, and CRP-CSF/CRP-COR. A similar strategy was used to generate Δcya/BpfD-Flag/BpfG-HA/CRP-His. All transformants were verified by PCR and DNA sequencing.
To replace the bpfA operon promoter (PbpfA) with the constitutive aacC1 promoter77, PbpfA and its flanking regions were amplified by the primers PbpfA-5F/PbpfA-3R and cloned into pK19mobsacB to yield an intermediate plasmid pK19-PbpfAUD. The intermediate plasmid was then used as a template to amplify the PbpfA-free part with the primers PbpfA-aacC1-KinF and PbpfA-aacC1-KinR, yielding a linearized plasmid pK19-UD. The aacC1 promoter region (PaacC1) was amplified using the plasmid pUCGm as a template with the primers PaacC1-F and PaacC1-R. Then, the PaacC1 was ligated to pK19-UD to yield pK19-PaacC1-UD, which was conjugated into the WT. Thus, PaacC1 was inserted and replaced PbpfA by homologous recombination, yielding WT/PaacC1-bpfA. The transformant was verified by PCR and DNA sequencing.
In order to analyze whether cAMP influences the interaction between CRP and BpfD, crp-R84L-His10 was knocked into Δcrp/PaacC1-bpfA/BpfD-Flag/BpfG-HA to yield WT/PaacC1-bpfA/BpfD-Flag/BpfG-HA/CRP-R84L-His. The binding ability between CRP and BpfD was compared to CRP-R84L and BpfD using a Co-IP assay.
Biofilm microtiter plate assay
Overnight cultured LB seed broth was diluted to OD600~0.01 using MM1 medium, and 100 μl of diluent was aliquoted into 96-well cell culture plates (NEST, China). When necessary, 1 mM exogenous adenosine 3′, 5′-cyclic monophosphate sodium salt monohydrate (cAMP) (Sigma-Aldrich, USA) was added into MM1 medium. The 96-well cell culture plates were statically incubated at 30 °C for different times. The biofilm assay was performed based on a crystal violet dyeing method51. Specifically, after incubation, the planktonic cells were removed and the surface attached cells were washed twice using deionized water. Each well was added with 150 μl of 1% crystal violet solution and incubated for 15 min. The crystal violet solution was removed and the wells were washed twice with 200 μl of deionized water. Then, 200 ul of 95% ethanol solution was added to each well, and the absorbance at 570 nm was measured using a microplate reader (ELx800, BioTek, USA) to determine the biofilm biomass.
RNA extraction and real-time RT-PCR (qRT-PCR) assay
S. putrefaciens CN32 cells grown in 96-well plates were harvested at appropriate time points for RNA extraction using the TRIzol method. Next, 2 μg total RNA was reverse transcribed to cDNA following the manufacturer’s protocol (Promega, USA), and cDNA was used as template for the qRT-PCR analysis. The qRT-PCR assay was performed using the Power SYBRTM Green PCR mix (Applied Biosystems, USA) and analyzed using the BioRad CFX96 Touch System. The PCR program included a pre-denaturation step at 95 °C for 10 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s; the fluorescence was measured at the end of each cycle. The primers used in qRT-PCR analysis were listed in Supplementary Table 3, and the 16S rRNA gene was selected as an internal control. All of the experiments were performed at least three times.
Protein purification
To purify His6-CRP and His6-CRP-R84L, pET28a-CRP and pET28a-CRP-R84L were constructed and introduced into E. coli BL21 (DE3) cells, which were cultured at 37 °C in LB medium to an OD600~0.6 and were induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 24 h at 16 °C, and the His6-CRP and His6-CRP-R84L proteins were purified by Ni-agarose resin (CoWin Biosciences, China) according to the manufacturer’s protocol.
To purify GST-BpfD-In (intracellular domains), pGEX-4T-1-BpfD-In was constructed and introduced into E. coli BL21 (DE3) cells, which were cultured at 37 °C in LB medium to an OD600~0.6. The cells were then inducted with 0.4 mM IPTG for 24 h at 16 °C, and the GST-BpfD intracellular domains were purified by Glutathione-Sepharose resin (Solarbio Life Sciences, China) according to the manufacturer’s protocol. Meanwhile, the GST protein was purified.
To purify MBP-1291 and MBP-1291(GGAAF), pMAL-c2x-1291 and pMAL-c2x-1291-GGAAF, were constructed and introduced into E. coli BL21 (DE3) cells, respectively, which were cultured at 37 °C in LB medium to an OD600~0.8 and were induced with 0.6 mM IPTG for 20 h at 16 °C, and the harvested cells were resuspended in MBP binding buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA). Proteins were purified with PurKineTM MBP-tag dextrin resin (Abbkine Scientific, China) using elution buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 10 mM maltose).
Electrophoretic mobility shift assay (EMSA)
To perform EMSA experiment, the DNA probe bpfA-pro covered the bpfA promoter region from –219 bp to +11 bp relative to the bpfA start codon was amplified, and the purified probe was labeled with digoxigenin (DIG) using the DIG Gel Shift Kit, 2nd Generation (Roche, USA). EMSA was performed according to the manufacturer’s protocol. When necessary, 1 μM cAMP was added. The primers used to amplify the EMSA probes are listed in Supplementary Table 3.
cAMP measurement
The intracellular cAMP concentration was measured using an established method78. Specifically, S. putrefaciens CN32 cells were cultured in 96-well plates. When necessary, 1 mM exogenous cAMP (Sigma-Aldrich, USA) was added to MM1 medium. Cells were harvested at 30 h by centrifugation at 13,000 g for 2 min at 4 °C and washed twice with pre-cooled phosphate-buffered saline (PBS) buffer. Then, the cell samples were divided into two parts: one part was used to determine the total protein concentration using a Quick Start Bradford 1×dye reagent (BioRad, USA), and the other was acetylated following the manufacturer’s protocol and used to measure the intracellular cAMP concentration with a Cyclic AMP ELISA Kit (Cayman Chemical, USA). The intracellular cAMP concentrations were converted to picomoles per milligram of protein.
c-di-GMP measurement
S. putrefaciens CN32 cells grown in 96-well plate were harvested at 30 h by centrifugation at 13,000 g for 2 min at 4 °C and washed twice with precooled PBS buffer. Then, the cell samples were divided into two parts: one part was used to determine the total protein concentration using a Quick Start Bradford 1×dye reagent (BioRad, USA); the other was lysed by B-PER Bacterial Protein Extraction Reagent (Thermo Fisher Scientific, USA), incubated at room temperature for 10 min, and then centrifuged at 13,000 g for 5 min. The liquid supernatant was used to measure the intracellular c-di-GMP concentration using a Cyclic di-GMP ELISA Kit (Cayman Chemical, USA). The intracellular c-di-GMP concentrations were converted to picomoles per milligram of protein.
DGC activity assays in vitro
The DGC activity was analyzed according to an established method79. Specifically, the protein Sputcn32_1291 and GTP were dissolved using the reaction buffer (50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl) to 10 μM and 300 μM, respectively. Then, 100 μl of Sputcn32_1291 solution was mixed with 300 μl of GTP solution, and incubated at 30 °C for 5 h. Subsequently, the reaction mixture was filtered through a 0.22 μm filter, and the filtrates were analyzed using a reversed-phase HPLC (EClassical 3100 HPLC system, Elite, China) equipped with a C18 column (Supersil ODS2, 5 μm, 4.6 × 200 mm, Elite, China). Buffer A (100 mM KH2PO4, 4 mM tetrabutyl ammonium hydrogen sulfate, pH 5.9) and buffer B (75% buffer A, 25% methanol) were used for product separation in a gradient program (minute and buffer B percentage): 0.0, 0%; 2.5, 0%; 5.0, 30%; 10.0, 60%; 14.0, 100%; 21.0, 100%; 22.0, 50%; 23.0, 0% and 30.0, 0% at 40 °C with a flow rate of 0.7 ml min−1. Nucleotides were detected at a wavelength of 254 nm.
BpfA localization assay
The cells of different strains grown in 96-well plates were harvested and adjusted to the same cell concentration. Briefly, 30 ml of adjusted cells was harvested by centrifugation at 13,000 g for 5 min at 4 °C, resuspended in 150 μl PBS, and mixed with 150 μl PBS containing 8 mg ml−1 lysozyme (Solarbio Life Sciences, China). The obtained cell suspensions were incubated at 37 °C for 15 min, and then centrifuged again (13,000 g, 4 °C, 5 min). Finally, the BpfA-containing supernatant fraction was assayed by western blotting and dot blotting.
Dot blotting
Dot blotting was performed using established method80,81. Specifically, the 10 μl sample of BpfA on the cell surface from different strains was dropped on a PVDF membrane (Roche Diagnostics, Germany), dried at 37 °C for 60 min, and incubated at 4 °C overnight. Flag-tagged BpfA was detected by western blotting analysis.
GST pull-down experiment
Proteins used for GST pull-down assay were heterologously expressed and purified. His6-CRP, His6-CRP-R84L, GST-BpfD-In, and GST protein were dialyzed with PBS-glycerol buffer (PBS buffer containing 20% of glycerol). Equal amounts of GST-tagged and His6-tagged proteins were mixed, and 20 μM cAMP was supplied if necessary. Pull-down buffer (50 mM Na2HPO4/NaH2PO4, pH 7.8, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40) was added to a final volume of 1 ml. Subsequently, 50 μl pre-balanced Glutathione-Sepharose Resin (Solarbio Life Sciences, China) was supplied to each volume. Following a 2 h incubation with generous rotation at 4 °C, the Glutathione-Sepharose Resin was washed five times with pull-down buffer, and 20 μM cAMP was supplied if necessary. The bound proteins were analyzed by western blotting.
Co-immunoprecipitation (Co-IP) assay
Co-IP was performed to verify the interaction between Flag-tagged BpfD and His-tagged CRP and the interaction between Flag-tagged BpfD and HA-tagged BpfG. Strains grown in 96-well plates at 30 h were harvested, washed once with PBS, and resuspended in ice-cold lysis buffer (45 mM HEPES, pH 7.2, 10% glycerol, 0.2% NP-40, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, and 1×protease inhibitor cocktail [CoWin Biosciences, China]) for 30 min. The cell lysates were clarified by centrifugation at 4 °C, 13,000 g for 20 min. The total protein concentration in the supernatant was determined by a Quick Start Bradford 1×dye reagent (BioRad, USA) and adjusted to 2 mg ml−1. The supernatant was incubated with monoclonal anti-Flag M2 antibody produced in mouse (Sigma-Aldrich, USA) and GammaBind G Sepharose beads (GE Healthcare, USA) on a rotary shaker at 4 °C for 2 h. The IgG from mouse serum (Sigma-Aldrich, USA) was used as negative control. Then, the protein-bead complexes were washed using lysis buffer for three times. When necessary, 5 μM c-di-GMP was added to the lysis buffer to perform interaction analysis between Flag-tagged BpfD and HA-tagged BpfG64, and 5 μM cAMP was added to the lysis buffer to perform interaction analysis between Flag-tagged BpfD and His-tagged CRP. Then, the bound protein complexes were eluted from beads using sample loading buffer and analyzed by western blotting.
Biotinylated cAMP pull-down assay
The binding between cAMP and CRP and CRP-R84L was analyzed using Dynabeads™ M-280 Streptavidin (Thermo Fisher Scientific, USA). The streptavidin magnetic beads were washed by TTBS (0.1 M Tris-HCl, pH 8.0, 0.9% (w/v) NaCl, 0.1% (v/v) Tween-20) before use. 2 nmol Biotin-cAMP conjugate (AAT Bioquest, USA) and equal amount of protein (His6-CRP or His6-CRP-R84L) were incubated in reaction buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM DTT) at 30 °C for 30 min. 2 nmol cAMP (Sigma-Aldrich, USA) or Biotin (Sigma-Aldrich, USA) were incubated with protein as controls. Half of the mixture was kept as input, while the other half was combined with the washed beads and incubated for 1.5 h at room temperature on a rotary shaker. The beads were collected using a magnetic stand and the supernatant was removed. After washing the beads using TTBS for three times, the beads were added with sample loading buffer and boiled to elute the bound protein, and the protein was then analyzed by western blotting.
Extraction of intracellular total proteins
In order to analyze the intracellular protein levels of Sputcn32_1291, Sputcn32_3328, and BpfA, the total proteins of S. putrefaciens CN32 cells collected from 96-well plates were extracted using B-PER Bacterial Protein Extraction Reagent (Thermo Fisher Scientific, USA); and the samples were adjusted to an equal amount of protein for western blotting assay.
Western blotting
The protein samples were separated by SDS-PAGE, and then transferred onto a PVDF membrane (Roche Diagnostics, Germany). After blocking with skim milk (5% in TBST), the membranes were incubated with primary antibody (Monoclonal anti-Flag M2 antibody produced in mouse, Sigma-Aldrich, USA; Anti HA-Tag mouse monoclonal antibody, Anti His-Tag mouse monoclonal antibody and Anti GST-Tag mouse monoclonal antibody were purchased from CoWin Biosciences, China) against the target protein was supplied, followed by incubation with Goat anti-mouse IgG HRP conjugated secondary antibody (CoWin Biosciences, China), or Goat Anti-Mouse IgG-Fc HRP conjugated secondary antibody (Sino Biological, China) for Co-IP experiments. The target protein was then detected using the eECL Western Blotting Kit (CoWin Biosciences, China) according to the manufacturer’s protocol.
Microscale thermophoresis (MST) assay
Microscale thermophoresis (MST) was performed to qualitatively analyze the interaction between BpfD-In and CRP. Proteins were heterologously expressed and purified. His6-CRP was dialyzed with PBS-glycerol buffer. GST and GST-BpfD-In were freeze-dried after dialysis with deionized water. Subsequently, 100 μl of 10 μM His6-CRP was labeled with NHS NT-647 dye using the Red-NHS 2nd Generation Labeling Kit (NanoTemper, Germany), which was eluted with a reaction buffer (PBS-glycerol buffer: Pull-down buffer = 1:1). Next, 10 μl of labeled His6-CRP was mixed with 10 μl of 5 μM GST or GST-BpfD-In or with the same volume of reaction buffer, respectively. Finally, samples were loaded into capillaries and analyzed using a Monolith Instrument NT.115 (NanoTemper, Germany) in Binding Check mode. Both the LED power and MST power were set to 20%.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism (version 9.0.0). All experiments were performed at least three independent times. Data are presented as a mean ± SD (standard deviation). Statistical significance was determined using two-sided Student’s t test. p values are reported using the following symbolic representation: NS (No significance) p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Professor Guoliang Qian for gifting the strain E. coli MG1655. This study is supported by National Natural Science Foundation of China (31970036, 31900401 and 31800020), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB180001, 20KJA180007); Natural Science Foundation of Jiangsu Province (BK20181009, BK20210920), Natural Science Foundation of Xuzhou city (KC19196), Six Talent Peaks Project of Jiangsu Province (JNHB-103), Priority Academic Program Development of Jiangsu Higher Education Institutions.
Source data
Author contributions
C.L., D.S., and W.J.L. planed and designed researches. C.L., Y.C., X.G.Z., Y.R.R., and J.R.Z. carried out the mutant constructions and biofilm assays. C.L., W.J.L., and D.S. performed blotting experiments, c-di-GMP and cAMP measurements. J.W.L. and D.S. carried out MST assay. C.L. and D.S. analyzed the data. W.J.L., C.L., and D.S. wrote the paper and all authors revised it.
Peer review
Peer review information
Nature Communications thanks Jason Smith and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Source Data are provided with this paper. All the data supporting this study are available in the main article, Supplementary Information files, Source Data file, or from the corresponding authors upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cong Liu, Di Sun
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-29240-5.
References
- 1.Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc. Natl Acad. Sci. USA. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunasekara SM, et al. Directed evolution of the Escherichia coli cAMP receptor protein at the cAMP pocket. J. Biol. Chem. 2015;290:26587–26596. doi: 10.1074/jbc.M115.678474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gronenborn AM, Sandulache R, Gärtner S, Clore GM. Mutations in the cyclic AMP binding site of the cyclic AMP receptor protein of Escherichia coli. Biochem. J. 1988;253:801–807. doi: 10.1042/bj2530801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youn H, Kerby RL, Conrad M, Roberts GP. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson CL, et al. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benoff B, et al. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 7.Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase α subunits. J. Mol. Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 8.Valentin‐Hansen P, Søgaard‐Andersen L, Pedersen H. A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol. Microbiol. 1996;20:461–466. doi: 10.1046/j.1365-2958.1996.5341056.x. [DOI] [PubMed] [Google Scholar]
- 9.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 10.Skorupski K, Taylor RK. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl Acad. Sci. USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CM, Ahn YJ, Kim SJ, Yoon DH, Shin SH. Temperature change induces the expression of vuuA encoding vulnibactin receptor and crp encoding cyclic AMP receptor protein in Vibrio vulnificus. Curr. Microbiol. 2016;73:54–64. doi: 10.1007/s00284-016-1026-8. [DOI] [PubMed] [Google Scholar]
- 12.Ou Q, et al. Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J. Med. Microbiol. 2017;66:1–7. doi: 10.1099/jmm.0.000391. [DOI] [PubMed] [Google Scholar]
- 13.Ritzert JT, Minasov G, Embry R, Schipma MJ, Satchell KJ. The cyclic AMP receptor protein regulates quorum sensing and global gene expression in Yersinia pestis during planktonic growth and growth in biofilms. mBio. 2019;10:e02613–e02619. doi: 10.1128/mBio.02613-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hufnagel DA, et al. The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J. Bacteriol. 2016;198:3329–3334. doi: 10.1128/JB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahramanoglou C, et al. Genomic mapping of cAMP receptor protein (CRPMt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42:8320–8329. doi: 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knapp GS, et al. Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res. 2015;43:5377–5393. doi: 10.1093/nar/gkv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gárate, F. et al. cAMP is an allosteric modulator of DNA-binding specificity in the cAMP receptor protein from Mycobacterium tuberculosis. J. Biolog. Chem. 296, 100480 (2021). [DOI] [PMC free article] [PubMed]
- 19.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 20.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 21.Saunders SH, et al. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell. 2020;182:919–932. e919. doi: 10.1016/j.cell.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katharios-Lanwermeyer S, Whitfield GB, Howell PL, O’Toole GA. Pseudomonas aeruginosa uses c-di-GMP phosphodiesterases RmcA and MorA to regulate biofilm maintenance. mBio. 2021;12:e03384–03320. doi: 10.1128/mBio.03384-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 26.Hengge R. Linking bacterial growth, survival, and multicellularity-small signaling molecules as triggers and drivers. Curr. Opin. Microbiol. 2020;55:57–66. doi: 10.1016/j.mib.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Peltier J, et al. Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease-mediated cleavage. J. Biol. Chem. 2015;290:24453–24469. doi: 10.1074/jbc.M115.665091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hun Yoon S, Waters CM. The ever-expanding world of bacterial cyclic oligonucleotide second messengers. Curr. Opin. Microbiol. 2021;60:96–103. doi: 10.1016/j.mib.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee, P., Sahoo, P. K., Adhikary, A., Ruhal, R. & Jain, D. Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa. Mol. Aspects Med. 81, 101001 (2021). [DOI] [PubMed]
- 30.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 31.Dahlstrom KM, O’Toole GA. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu. Rev. Microbiol. 2017;71:179–195. doi: 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarenko O, et al. More than enzymes that make or break cyclic di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio. 2017;8:e01639–01617. doi: 10.1128/mBio.01639-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LH, et al. Cyclic di-GMP-dependent signaling pathways in the pathogenic Firmicute Listeria monocytogenes. PLoS Pathog. 2014;10:e1004301. doi: 10.1371/journal.ppat.1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengge, R. High-specificity local and global c-di-GMP signaling. Trends Microbiol. 29, 993–1003 (2021). [DOI] [PubMed]
- 35.Chen G, et al. The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J. 2020;39:e103412. doi: 10.15252/embj.2019103412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt JH, et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio. 2010;1:e00183–00110. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S, Dingemans J, Gowett M, Sauer K. Glucose-6-phosphate acts as an extracellular signal of SagS to modulate Pseudomonas aeruginosa c-di-GMP levels, attachment, and biofilm formation. mSphere. 2021;6:e01231–01220. doi: 10.1128/mSphere.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee D, et al. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife. 2014;3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floyd KA, et al. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-15331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kunz S, et al. Cyclic di-GMP signaling in Bacillus subtilis is governed by direct interactions of diguanylate cyclases and cognate receptors. mBio. 2020;11:e03122–03119. doi: 10.1128/mBio.03122-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins AJ, Smith TJ, Sondermann H, O’Toole GA. From input to output: the lap/c-di-GMP biofilm regulatory circuit. Annu. Rev. Microbiol. 2020;74:607–631. doi: 10.1146/annurev-micro-011520-094214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martínez-Méndez, R., Camacho-Hernández, D. A., Sulvarán-Guel, E. & Zamorano-Sánchez, D. A trigger phosphodiesterase modulates the global c-di-GMP pool, motility and biofilm formation in Vibrio parahaemolyticus. J. Bacteriol. 203, e0004621 (2021). [DOI] [PMC free article] [PubMed]
- 44.Heidelberg JF, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 45.Fredrickson JK, et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 46.Khalid A, Arshad M, Crowley DE. Decolorization of azo dyes by Shewanella sp. under saline conditions. Appl. Microbiol. Biotechnol. 2008;79:1053–1059. doi: 10.1007/s00253-008-1498-y. [DOI] [PubMed] [Google Scholar]
- 47.Liu W, et al. The cyclic AMP receptor protein, Crp, is required for the decolorization of acid yellow 36 in Shewanella putrefaciens CN32. Front. Microbiol. 2020;11:2783. doi: 10.3389/fmicb.2020.596372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee M, Zaiden N, Teng A, Hu Y, Cao B. Shewanella biofilm development and engineering for environmental and bioenergy applications. Curr. Opin. Chem. Biol. 2020;59:84–92. doi: 10.1016/j.cbpa.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Theunissen S, et al. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res. Microbiol. 2010;161:144–152. doi: 10.1016/j.resmic.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Wu C, et al. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci. Rep. 2013;3:1–7. doi: 10.1038/srep01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, et al. Sodium lactate negatively regulates Shewanella putrefaciens CN32 biofilm formation via a three-component regulatory system (LrbS-LrbA-LrbR) Appl. Environ. Microbiol. 2017;83:e00712–e00717. doi: 10.1128/AEM.00712-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng YY, et al. FlrA represses transcription of the biofilm-associated bpfA operon in Shewanella putrefaciens. Appl. Environ. Microbiol. 2017;83:e02410–e02416. doi: 10.1128/AEM.02410-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theunissen S, et al. The agglutination protein AggA from Shewanella oneidensis MR-1 is a TolC-like protein and forms active channels in vitro. Biochem. Biophys. Res. Commun. 2009;386:380–385. doi: 10.1016/j.bbrc.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 54.De Windt W, et al. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology. 2006;152:721–729. doi: 10.1099/mic.0.28204-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou G, Yuan J, Gao H. Regulation of biofilm formation by BpfA, BpfD, and BpfG in Shewanella oneidensis. Front. Microbiol. 2015;6:790. doi: 10.3389/fmicb.2015.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schniederberend M, et al. Modulation of flagellar rotation in surface-attached bacteria: a pathway for rapid surface-sensing after flagellar attachment. PLoS Pathog. 2019;15:e1008149. doi: 10.1371/journal.ppat.1008149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 58.West S, Sample AK, Runyen-Janecky LJ. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin CT, et al. CRP-cyclic AMP regulates the expression of type 3 fimbriae via cyclic di-GMP in Klebsiella pneumoniae. PLoS ONE. 2016;11:e0162884. doi: 10.1371/journal.pone.0162884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almblad H, et al. High levels of cAMP inhibit Pseudomonas aeruginosa biofilm formation through reduction of the c-di-GMP content. Microbiology. 2019;165:324–333. doi: 10.1099/mic.0.000772. [DOI] [PubMed] [Google Scholar]
- 61.Fong JC, Yildiz FH. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Sun D, Zhu J, Liu J, Liu W. The regulation of bacterial biofilm formation by cAMP-CRP: a mini-review. Front. Microbiol. 2020;11:802. doi: 10.3389/fmicb.2020.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu G, et al. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res. 2018;46:9276–9288. doi: 10.1093/nar/gky803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giacalone D, et al. Ligand-mediated biofilm formation via enhanced physical interaction between a diguanylate cyclase and its receptor. mBio. 2018;9:e01254–01218. doi: 10.1128/mBio.01254-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreira RN, et al. BolA is required for the accurate regulation of c-di-GMP, a central player in biofilm formation. mBio. 2017;8:e00443–00417. doi: 10.1128/mBio.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rumbaugh KP, Sauer K. Biofilm dispersion. Nat. Rev. Microbiol. 2020;18:571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hengge R. Crosstalking second messengers. Nat. Microbiol. 2021;6:9–10. doi: 10.1038/s41564-020-00842-3. [DOI] [PubMed] [Google Scholar]
- 69.Kalia D, et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP,(p) ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 70.Shyp V, et al. Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus. Nat. Microbiol. 2021;6:59–72. doi: 10.1038/s41564-020-00809-4. [DOI] [PubMed] [Google Scholar]
- 71.Kim C, et al. Antiinflammatory cAMP signaling and cell migration genes co-opted by the anthrax bacillus. Proc. Natl Acad. Sci. USA. 2008;105:6150–6155. doi: 10.1073/pnas.0800105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutrina SL, et al. The quantity and distribution of biofilm growth of Escherichia coli strain ATCC 9723 depends on the carbon/energy source. Microbiology. 2019;165:47–64. doi: 10.1099/mic.0.000745. [DOI] [PubMed] [Google Scholar]
- 73.Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 74.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 75.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 77.Wohlleben W, et al. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC (3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 1989;217:202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- 78.Almblad H, et al. The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic di-GMP. J. Bacteriol. 2015;197:2190–2200. doi: 10.1128/JB.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monds RD, Newell PD, Gross RH, O’Toole GA. Phosphate‐dependent modulation of c‐di‐GMP levels regulates Pseudomonas fluorescens Pf0‐1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 81.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3′, 5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl Acad. Sci. USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source Data are provided with this paper. All the data supporting this study are available in the main article, Supplementary Information files, Source Data file, or from the corresponding authors upon request. Source data are provided with this paper.