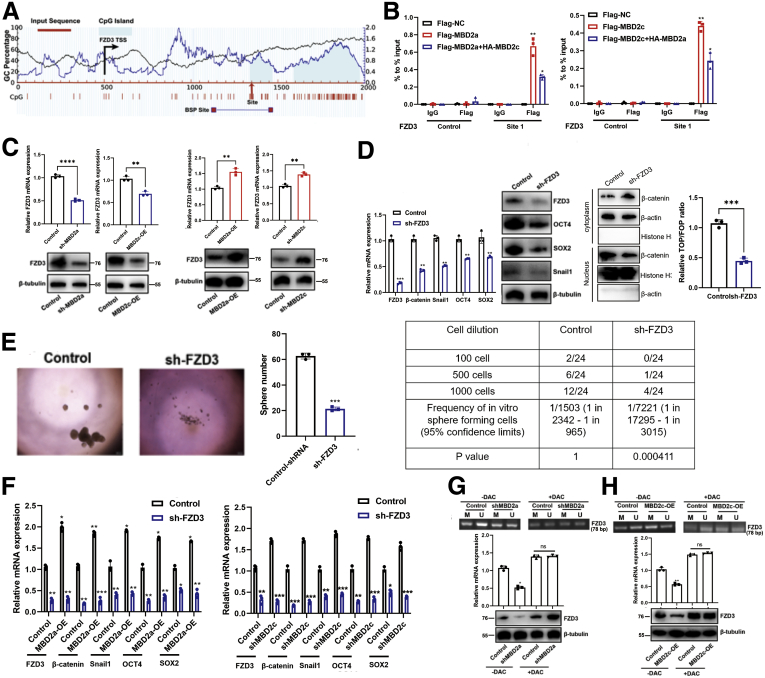
Figure 12.
MBD2a and MBD2c competitive binding to the CpG islands in the FZD3 promoter.A, Bioinformatic analysis of FZD3 gene. Location of CpG islands (light blue), TSS (black), qPCR sites (site 1 with red arrow), and BSP-amplified regions were indicated in this graph. Indicated positions were relative to the TSS (+1). BSP, Bisulfite sequencing PCR; TSS, transcription start site. B, ChIP experiments were performed using IgG or Flag antibody in indicated MHCC97H cells. The occupancy of the predicted DNA binding site for MBD2a or MBD2c within CpG islands of the FZD2 promoter assessed using qPCR. Data are presented as the mean (± standard deviation) of 3 independent experiments. ∗P < .05 as compared with the corresponding Flag-EV+HA-EV samples. C, qPCR and immunoblot analyses of FZD3 expression in MHCC97H cells stably expressing shRNAs targeting MBD2a, or overexpression of MBD2c. ∗P < .05; ∗∗P < .01 as compared with the corresponding control or EV control samples. D, qPCR and immunoblot analyses of β-catenin, FZD3, OCT4, SOX2, and Snail1 expression in MHCC97H cells stably expressing NC or FZD3 shRNAs. ∗∗P < .01; ∗∗∗P < .001 as compared with control samples. E, Sphere formation and limiting dilution assays for shRNA targeting FZD3 in MHCC97H cells. Data were from 3 independent experiments, ∗∗∗P < .001 by the Student t test. F, MHCC97H cells stably expressing EV, MBD2a (left panel), control NTC, MBD2c shRNAs (right panel) were further transfected with viruses expressing control NC, or FZD3 shRNAs, then the expression of FZD3, OCT4, SOX2, β-catenin, and Snail1 were measured by qPCR. ∗P < .05 as compared with the corresponding controls. G, MHCC97H cells stably expressing empty virus (EV), MBD2c (H), or control, MBD2a shRNAs (G) were further treated without or with DAC (20 μM) for 48 hours, followed by MSP detection of the methylation level of CpG island in FZD3 promoter and qPCR and immunoblot analyses (bottom panel) of the mRNA and protein expression of FZD3. PCR products are amplified with primers that recognize the methylated (M) or unmethylated sequences (U). ∗P < .05 as compared with the corresponding EV or control samples. ns, Not significant. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test.