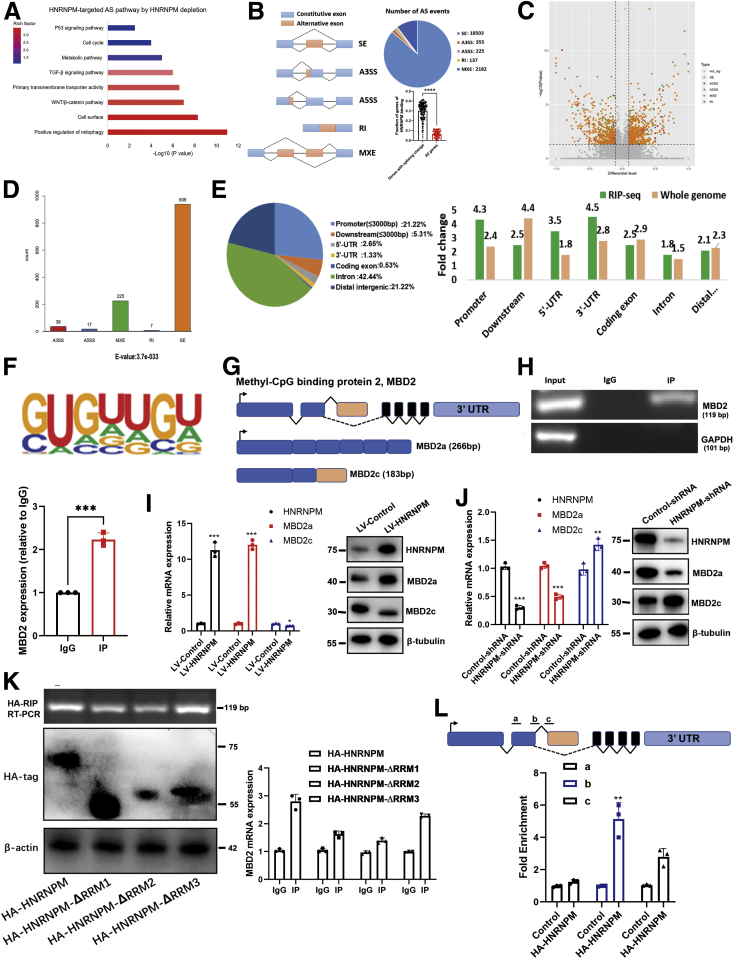
Figure 8.
The genome-wide landscape and global alternative splicing of HNRNPM.A, Kyoto Encyclopedia of Genes and Genomes analysis of HNRNPM-targeted splicing events. B, Quantification of the different AS events regulated by HNRNPM. A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site; MXE, mutually exclusive exon; RI, retained intron; SE, skipped exon. ∗∗∗P < .001 by the Student t test. C-D, The quantification of significant AS events regulated by HNRNPM (P < .05). E, HNRNPM-RIP-seq peaks were enriched in 5′UTR, promoter and 3′ UTR. All RIP-seq peaks were categorized according to the distribution on different genomic elements andcompared with the genomic background. F, De novo motif analysis identifying GU-repeat motif as the only enriched motif within the top HNRNPM RIP-seqpeaks. G, Schematic diagram of MBD2 molecular model. H, The RIP experiment showed HNRNPM directly binded with MBD2. I, The shift of MBD2a and MBD2c between HNRNPM overexpressed stably transduced and control MHCC97H cells. J, The shift of MBD2a and MBD2c between HNRNPM shRNA stably transduced and control MHCC97H cells. K, The RMMs of HNRNPM bind to MBD2 by RIP experiments. L, The potential binding of HNRNPM to MBD2 pre-mRNA by CLIP assay.