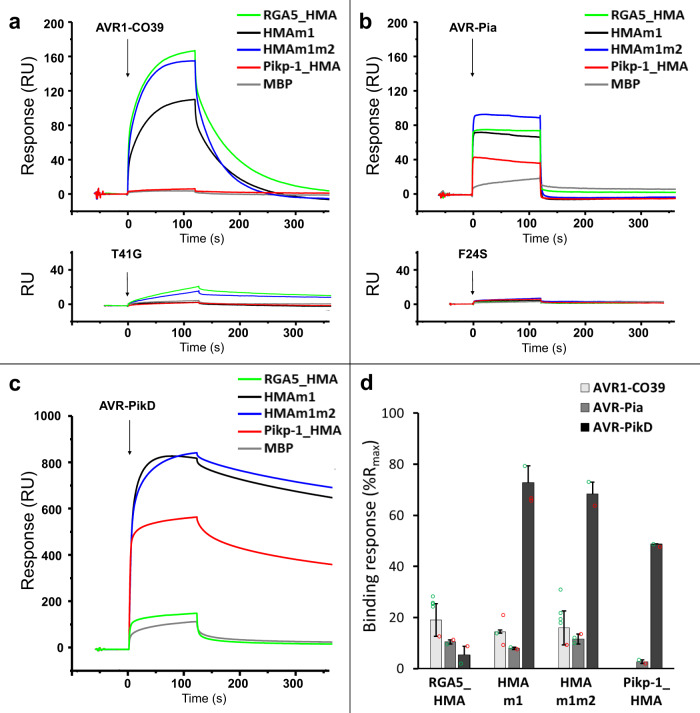
Fig. 3. Engineered RGA5_HMAs bind strongly to AVR-PikD in vitro.
a–c AVR1-CO39 (a), AVR-Pia (b), and AVR-PikD (c) were injected (black arrows) at 1 µM for 2 min on the different MBP:HMA fusion proteins captured by anti-MBP antibody immobilized on the chip. Superimposed sensorgrams are shown for wild-type RGA5_HMA (green), RGA5_HMAm1 (black), RGA5_HMAm1m2 (blue), Pikp-1_HMA (red), as well as for MBP alone (gray) that serves as a negative control. The binding curves obtained with the wild-type and an inactive variant of AVR1-CO39 (a) or AVR-Pia (b) are shown in the top and lower insets, respectively. d Comparison of the binding response (bound fraction) at 1 µM of AVR effectors, expressed as the percentage of the theoretical maximum response (%Rmax) normalized for the amount of MBP:HMA immobilized on the chip and corrected for the MBP-tag contribution. Bars and error bars represent the mean and average deviation calculated for the %Rmax values estimated from n = 2 independent experiments carried out on two different days (open green and red circles) wit n = 1 to n = 4 technical replicates per experiment. Two independently purified protein samples were used to test the binding of the AVR-PikD effector to the different HMAs.