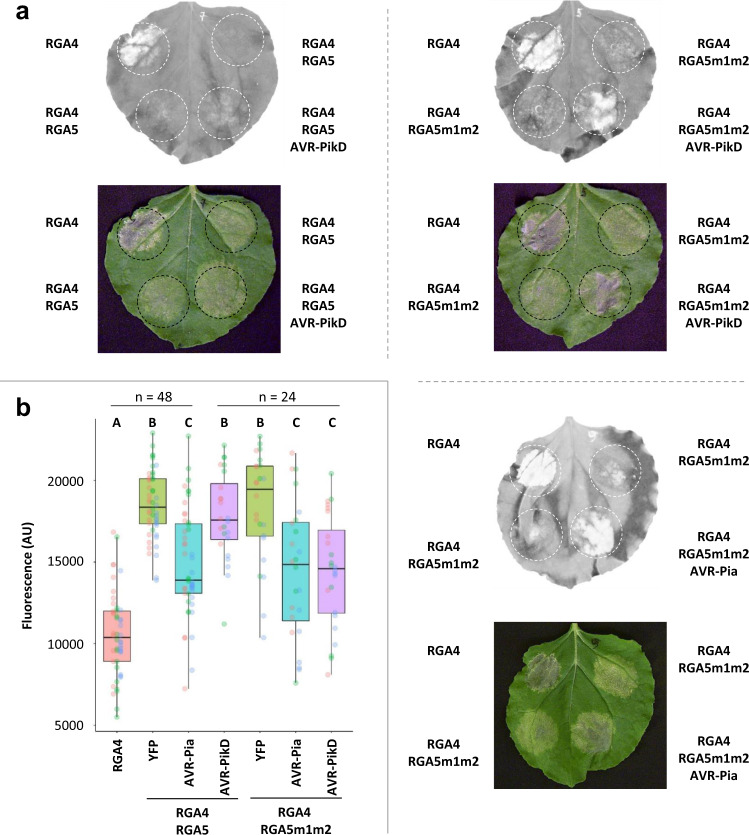
Fig. 7. The RGA5m1m2 mutant recognizes AVR-Pia and AVR-PikD in N. benthamiana.
a The indicated combinations of constructs were transiently expressed in N. benthamiana leaves. The AVR-PikD (without signal peptide) and RGA4 constructs carry an HA tag at their C-termini, while RGA5 and RGA5m1m2 are YFP-tagged at their N-termini and AVR-Pia (without signal peptide) is untagged. The auto-active RGA4 construct was used as a positive control for cell death induction. Cell death was visualized 5 days after infiltration. Greyscale pictures were taken using a fluorescence scanner with settings allowing visualization of the disappearance of red fluorescence due to cell death (white patches of dead cells). A picture of corresponding leaves is shown below. b Cell death was quantified by measuring fluorescence levels (arbitrary unit, AU) in the infiltrated areas using ImageJ34. The resulting data were plotted. The boxes represent the first quartile, median, and third quartile. The difference in fluorescence levels was assessed by an ANOVA followed by a Tukey HSD test. Groups with the same letter (a–c) are not significantly different at level 0.05. For each combination of constructs, all of the measurements are represented as dots with a distinct color (red, green, and blue) for each of the three biological replicates.