Figure 5.
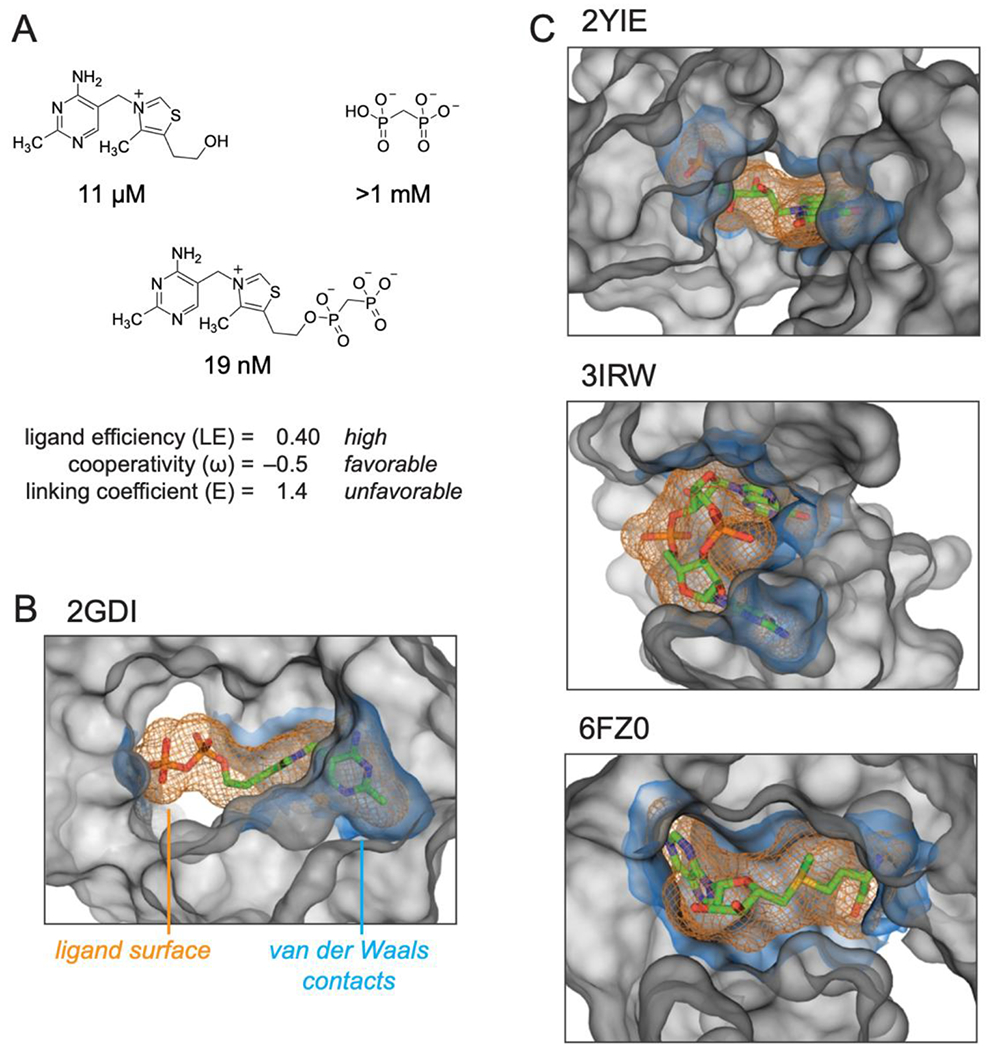
Summary of the structures and properties of thiamine and MDP fragments and the linked compound, TPPc, and comparison with representative RNA structures that interact with their ligands using well-defined sub-sites. (A) TPPc features. Ligand efficiency (LE) is equal to the binding energy per non-hydrogen ligand atom, LE = ΔG / N; ligand cooperativity, ω, is defined in Table 1; linking coefficient (E) was calculated as E = KL / (KT KP). (B) TPP riboswitch binding pocket. (C) Representative examples of riboswitch ligands that bind in defined sub-sties: FMN43, cyclic di-GMP44, and SAM-V riboswitches45. Ligands are shown as sticks, orange mesh shows ligand molecular envelope. RNA atoms within 6 Å of ligand are shown as a grey surface that delineates the topography of the binding pocket. The RNA surface within 2 Å of the ligand, equal to the van der-Waals contact distance57, is shown in blue. VDW surfaces were calculated using HOLLOW57 and were visualized in PyMOL (Schrodinger, LLC). PDB IDs for each structure are shown.
