Abstract
Recently, a novel variant p.Y314S in UQCRC1 has been implicated as pathogenic in Parkinson’s disease (PD). In the present study, we aimed to examine the association of UQCRC1 with PD in large cohorts of European origin. We examined common and rare genetic variation in UQCRC1 using genome-wide association study data from the International Parkinson Disease Genomics Consortium, including 14,671 cases and 17,667 controls, and whole-genome sequencing data from the Accelerating Medicines Partnership–Parkinson’s disease initiative, including 1647 patients with PD and 1050 controls. No common variants were consistently associated with PD, and a variety of burden analyses did not reveal an association between rare variants in UQCRC1 and PD. Therefore, our results do not support a major role for UQCRC1 in PD in the European population, and additional studies in other populations are warranted.
Keywords: Parkinson disease, Genetics, UQCRC1
1. Introduction
The genetics of Parkinson’s disease (PD) has been extensively studied over the last 20 years (Bandres-Ciga et al., 2020). A recent genome-wide association study (GWAS) identified 90 independent risk variants (Nalls et al., 2019); however, these variants explain less than 50% of the heritability of PD (Nalls et al., 2019), suggesting that other, unknown common and rare genetic variants affect the risk of PD. A novel variant p.Y314S in UQCRC1 encoding the ubiquinol-cytochrome c reductase core protein (UQCRC1) has been recently identified in 5 Taiwanese family members with parkinsonism by whole-exome sequencing (Lin et al., 2019). UQCRC1 is a mitochondrial protein and part of the respiratory chain III complex (Hoffman et al., 1993), which may play a role in mitochondrial respiration (Lin et al., 2019; Shan et al., 2019).
The purpose of this work is to examine the role of UQCRC1 in a large-scale European PD population using GWAS and whole-genome sequencing (WGS) data from the International Parkinson Disease Genomics Consortium (IPDGC) and Accelerating Medicines Partnership–Parkinson’s disease (AMP-PD) initiative.
2. Methods
The study populations included 14,671 patients with PD and 17,667 controls from IPDGC and 1647 patients with PD and 1050 controls from AMP-PD (https://amp-pd.org/). Quality control of IPDGC GWAS data was performed on both individual and variant levels as previously described (Nalls et al., 2019). Similar quality control procedures were performed in the AMP-PD WGS data, as described by AMP-PD (https://amp-pd.org/whole-genome-data). We extracted UQCRC1 genotyping data from both data sets using gene coordinates determined by UQCRC1 position (+/− 100kb): hg19: chr3:48,536,432–48,747,098; hg38: chr3:48,499,002–48,709,646. We used ANNOVAR (Wang et al., 2010) to annotate both data sets. PLINK 1.9 (Chang et al., 2015) was used for logistic regression (adjusted for age, sex, and first 10 principal components) to study the association between common UQCRC1 variants (with minor allele frequency [MAF] > 0.01) and PD in the IPDGC cohort. In the AMP-PD cohort, to study the burden of rare variants (defined as variants with MAF <0.03 in the current data), a variety of methods were applied, including sequence kernel association test and its optimized version, combined multivariate and collapsing, Zeggini and Madsen-Browning tests, as a part of the Rvtest package (Zhan et al., 2016). Bonferroni correction was applied to correct for multiple comparisons. All codes used in the present study are available at our GitHub at https://github.com/ipdgc/IPDGC-Trainees/blob/master/UQCRC1_IPDGC_trainee.md.
3. Results
Using the IPDGC GWAS data, we identified 140 common variants in the selected region (Fig. 1). None of the common variants annotated to UQCRC1 were associated with PD (Supplementary Table 1). In the AMP-PD WGS data, we identified 94 variants with MAF <0.03 within or close to UQCRC1 (Supplementary Table 2), including 9 nonsynonymous variants (Supplementary Table 2). We performed burden tests for 3 categories of variants: 1) all rare variants, 2) all rare coding variants, and 3) all rare nonsynonymous variants. None of the burden tests showed association between rare UQCRC1 variants and PD (Table 1). In both cohorts, we did not find the UQCRC1 p.Y314S variant described in the original study (Lin et al., 2019).
Fig. 1.
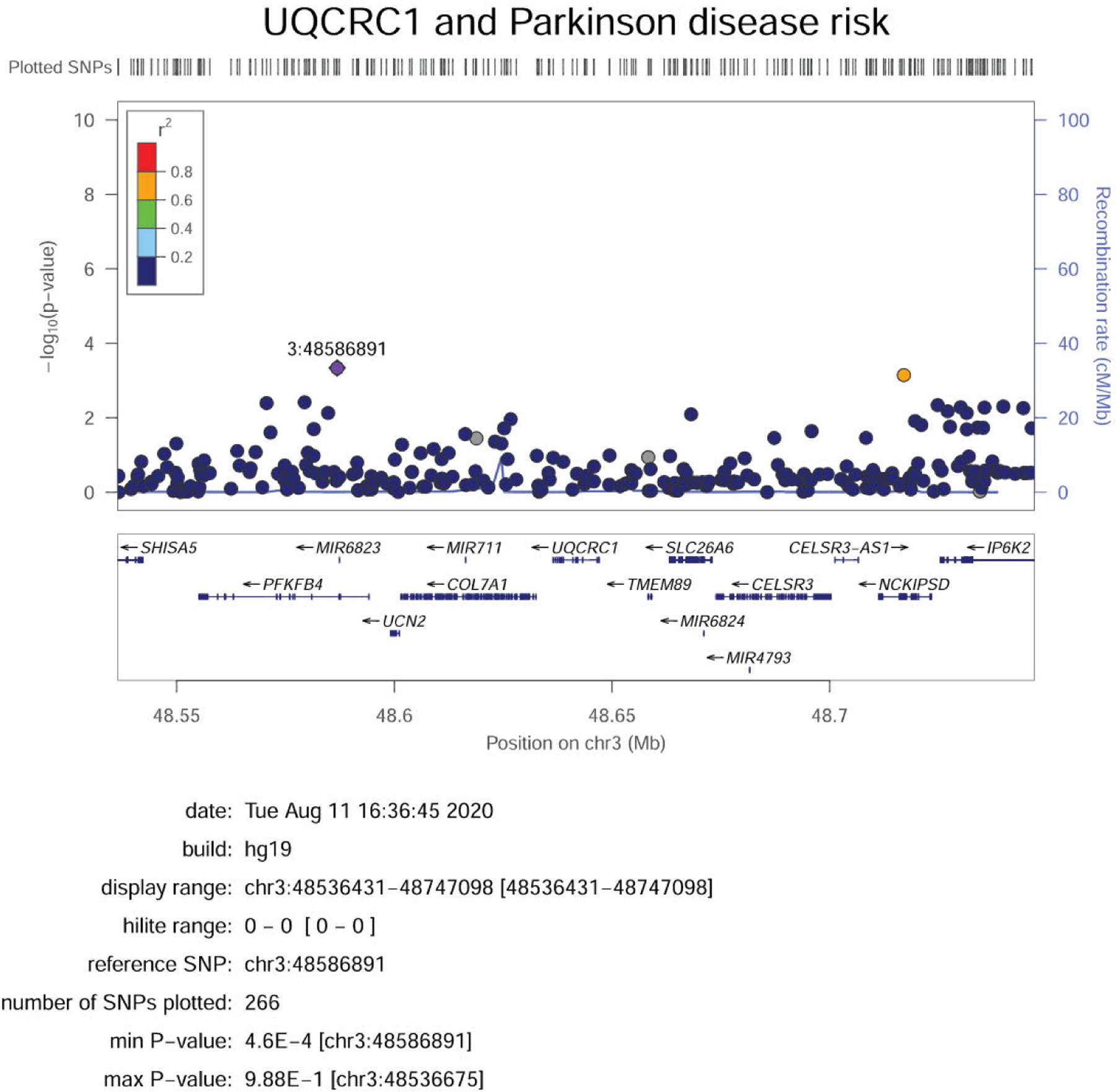
LocusZoom plot of IPDGC GWAS summary statistics showing variants with MAF more than 1% near the UQCRC1 gene. Abbreviations: GWAS, genome-wide association study; IPDGC, International Parkinson Disease Genomics Consortium; MAF, minor allele frequency.
Table 1.
Burden tests for UQCRC1 in the AMP-PD cohort
| Burden test | All variants | Coding variants | Nonsynonymous variants | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NumVar | NumPolyVar | p-value | NumVar | NumPolyVar | p-value | NumVar | NumPolyVar | p-value | |
| Zeggini | 73 | 54 | 0.562 | 18 | 13 | 0.619 | 12 | 9 | 0.721 |
| SKAT-O | 73 | 54 | 0.77 | 18 | 13 | 0.586 | 12 | 9 | 0.712 |
| SKAT | 73 | 54 | 0.865 | 18 | 13 | 0.476 | 12 | 9 | 0.594 |
| Madson-Browning | 73 | 54 | 0.487 | 18 | 13 | 0.046 | 12 | 9 | 0.109 |
| Fp | 73 | 54 | 0.712 | 18 | 13 | 0.489 | 12 | 9 | 0.898 |
| CMC | 73 | 54 | 0.596 | 18 | 13 | 0.629 | 12 | 9 | 0.731 |
Key: AMP-PD, Accelerating Medicines Partnership–Parkinson’s disease; CMC, combined multivariate and collapsing; NumPolyVar, number of polymorphic genotypes; NumVar, number of variants; SKAT, sequence kernel association test; SKAT-O, sequence kernel association test optimized version.
4. Discussion
In the present study, we performed a comprehensive analysis of common and rare variants in UQCRC1 using GWAS and WGS data from large cohorts of patients with PD and controls.
In the original study in which UQCRC1 variants were implicated, the authors described a family in which 5 carriers of UQCRC1 p.Y314S had late-onset levodopa responsive parkinsonism and axonal type sensorimotor polyneuropathy (Lin et al., 2019). In addition, autosomal dominant inheritance was demonstrated and there were no asymptomatic carriers of this variant, based on a family tree authors present (Lin et al., 2019). Recent replication study in an eastern Chinese population did not reveal association between UQCRC1 and PD (Lin et al., 2020).
It is possible that this variant is associated with a specific form of atypical parkinsonism, but not with typical PD. It has been shown that a number of genes previously reported as PD-associated (such as DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1) do not play a role in PD and thus should not be regarded as PD genes (Foo et al., 2014; Krüger et al., 2011; Lesage et al., 2010; Saini et al., 2020). Furthermore, other genes (e.g., ATP13A2, FBXO7) that are associated with atypical forms of parkinsonism are often cited as PD-associated genes (Dehay et al., 2012; Deng et al., 2015; Weissbach et al., 2019). It is important to properly define which genes are involved in typical PD and which genes are not, particularly in the era of targeted drug development.
Overall, we did not find any evidence to support an important role for UQCRC1 in patients with PD of European origin. However, additional studies in other populations are required to further study the potential role of UQCRC1 in PD, and we cannot rule out the possibility that very rare, specific UQCRC1 variants are associated with PD or with atypical forms of parkinsonism.
Supplementary Material
Acknowledgements
We thank the participants for contributing to the study. We would like to also thank all members of the International Parkinson Disease Genomics Consortium (IPDGC). For a complete overview of members, acknowledgements, and funding, please see http://pdgenetics.org/partners. Data used in the preparation of this article were obtained from the AMP PD Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org.” AMP-PD—a public-private partnership—is managed by the FNIH and funded by Celgene, GSK, the Michael J. Fox Foundation for Parkinson’s Research, the National Institute of Neurological Disorders and Stroke, Pfizer, Sanofi, and Verily. We would like to thank AMP-PD for the publicly available whole-genome sequencing data, including cohorts from the Fox Investigation for New Discovery of Biomarkers (BioFIND), the Parkinson’s Progression Markers Initiative (PPMI), and the Parkinson’s Disease Biomarkers Program (PDBP). BioFIND is sponsored by The Michael J. Fox Foundation for Parkinson’s Research (MJFF) with support from the National Institute for Neurological Disorders and Stroke (NINDS). For up-to-date information on the study, visit michaeljfox.org/biofind. PPMI—a public-private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including [list the full names of all of the PPMI funding partners found at www.ppmi-info.org/fundingpartners]. For up-to-date information on the study, visit www.ppmi-info.org. Parkinson’s Disease Biomarker Program (PDBP) consortium is supported by the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The BioFIND, PPMI, and PDBP Investigators have not participated in reviewing the data analysis or content of the manuscript. This work was financially supported by grants from the Michael J. Fox Foundation, the Canadian Consortium on Neurodegeneration in Aging (CCNA), the Canada First Research Excellence Fund (CFREF), awarded to McGill University for the Healthy Brains for Healthy Lives initiative (HBHL), and Parkinson Canada. This research was supported in part by the Intramural Research Program of the NIH, National institute on Aging. K.S. is supported by a postdoctoral fellowship from the Canada First Research Excellence Fund (CFREF), awarded to McGill University for the Healthy Brains for Healthy Lives initiative (HBHL). Z.G-O. is supported by the Fonds de recherche du Québec - Santé (FRQS) Chercheurs-boursiers award, in collaboration with Parkinson Quebec and by the Young Investigator Award by Parkinson Canada.
Footnotes
Disclosure statement
Z.G-O. has received consulting fees from Lysosomal Therapeutics Inc., Idorsia, Prevail Therapeutics, Denali, Ono Therapeutics, Neuron23, Handl Therapeutics, Deerfield and Inception Sciences (now Ventus). None of these companies were involved in any parts of preparing, drafting, and publishing this study. Other authors have no additional disclosures to report.
CRediT authorship contribution statement
Konstantin Senkevich: Conceptualization, Data curation, Formal analysis, Methodology, Visualization, Writing - original draft. Sara Bandres-Ciga: Data curation, Resources, Formal analysis, Software, Writing - review & editing. Ziv Gan-Or: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Visualization, Writing - review & editing. Lynne Krohn: Conceptualization, Methodology, Project administration, Resources, Supervision, Visualization, Writing - review & editing.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neurobiolaging.2020.10.030.
References
- Bandres-Ciga S, Diez-Fairen M, Kim JJ, Singleton AB, 2020. Genetics of Parkinson’s disease: an introspection of its journey towards precision medicine. Neurobiol. Dis 137, 104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Giga-science 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron M-H, Doudnikoff E, Vital A, Vila M, Klein C, Bezard E, 2012. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc. Natl. Acad. Sci 109, 9611–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Wu Y, Jankovic J, 2015. The EIF 4G1 gene and Parkinson’s disease. Acta Neurol. Scand 132, 73–78. [DOI] [PubMed] [Google Scholar]
- Foo JN, Liany H, Tan LC, Au W-L, Prakash K-M, Liu J, Tan E-K, 2014. DNAJ mutations are rare in Chinese Parkinson’s disease patients and controls. Neurobiol. Aging 35, 935. e931–935. e932. [DOI] [PubMed] [Google Scholar]
- Hoffman G, Lee S, Christiano A, Chung-Honet L, Cheng W, Katchman S, Uitto J, Greenspan D, 1993. Complete coding sequence, intron/exon organization, and chromosomal location of the gene for the core I protein of human ubiquinol-cytochrome c reductase. J. Biol. Chem 268, 21113–21119. [PubMed] [Google Scholar]
- Krüger R, Sharma M, Riess O, Gasser T, Van Broeckhoven C, Theuns J, Aasly J, Annesi G, Bentivoglio AR, Brice A, 2011. A large-scale genetic association study to evaluate the contribution of Omi/HtrA2 (PARK13) to Parkinson’s disease. Neurobiol. Aging 32, 548. e549–548. e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Condroyer C, Lohman E, Troiano A, Tison F, Viallet F, Damier P, Tranchant C, Vidhaillet M, Ouvrard-Hernandez A-M, 2010. Follow-up study of the GIGYF2 gene in French families with Parkinson’s disease. Neurobiol. Aging 31, 1069–1071. [DOI] [PubMed] [Google Scholar]
- Lin CH, Chen PL, Tai CH, Lin HI, Chen CS, Chen ML, Wu RM, 2019. A clinical and genetic study of early-onset and familial parkinsonism in taiwan: an integrated approach combining gene dosage analysis and next-generation sequencing. Mov Disord. 34, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZH, Zheng R, Ruan Y, Gao T, Jin CY, Xue NJ, Dong JX, Yan YP, Tian J, Pu JL, Zhang BR, 2020. The lack of association between ubiquinol-cytochrome c reductase core protein I (UQCRC1) variants and Parkinson’s disease in an eastern Chinese population. CNS Neurosci. Ther 26. 990–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simon-Sanchez J, Schulte C, Sharma M, Krohn L, Pihlstrom L, Siitonen A, Iwaki H, Leonard H, Faghri F, Gibbs JR, Hernandez DG, Scholz SW, Botia JA, Martinez M, Corvol JC, Lesage S, Jankovic J, Shulman LM, Sutherland M, Tienari P, Majamaa K, Toft M, Andreassen OA, Bangale T, Brice A, Yang J, Gan-Or Z, Gasser T, Heutink P, Shulman JM, Wood NW, Hinds DA, Hardy JA, Morris HR, Gratten J, Visscher PM, Graham RR, Singleton AB, Me Research, T., System Genomics of Parkinson’s Disease. C., International Parkinson’s Disease Genomics, C., 2019. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Rudakou U, Yu E, Ruskey J, Asayesh F, Laurent S, Spiegelman D, Fahn S, Waters C, Monchi O, 2020. Association of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with Parkinson’s disease. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Li J, Xu W, Li H, Zuo Z, 2019. Critical role of UQCRC1 in embryo survival, brain ischemic tolerance and normal cognition in mice. Cell Mol. Life Sci 76, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H, 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach A, Wittke C, Kasten M, Klein C, 2019. ‘Atypical’Parkinson’s Disease–Genetic, International Review of Neurobiology. Elsevier, pp. 207–235. https://www.sciencedirect.com/science/article/pii/S0074774219300972?via%3Dihub. (Accessed 2 August 2020). [DOI] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ, 2016. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32,1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


