Extended Data Fig. 8, Asparagine and glutamine can be utilized as sole nitrogen sources by most tested Bacteroidetes.
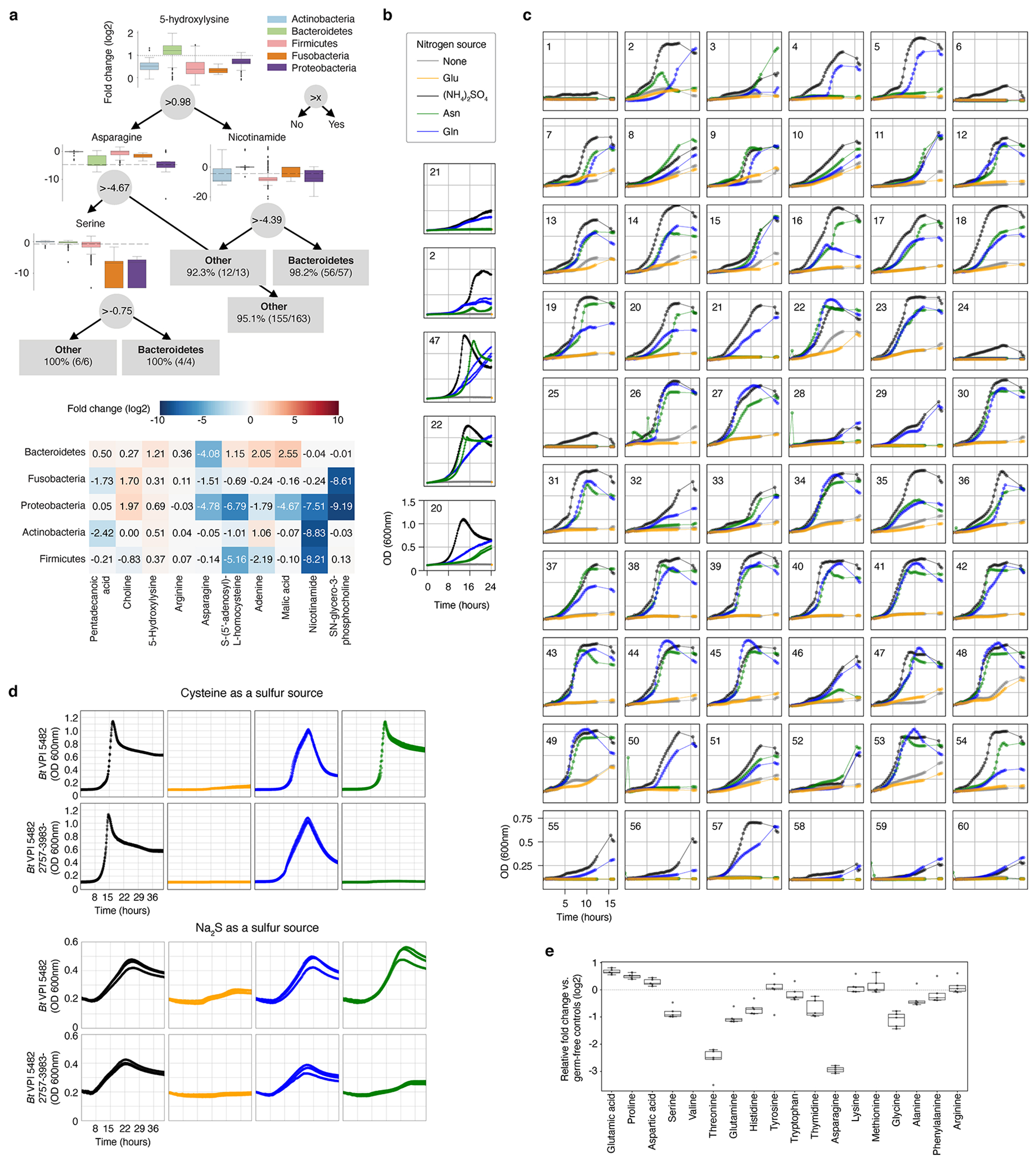
a, Top panel: An example decision tree from a forest that can differentiate Bacteroidetes vs. bacteria from the other four represented phyla with > 97% accuracy. For each decision node, phylum-level elevation and reduction based on metabolite levels are shown (relative fold change to bacterial media controls, log2 transformed). Actinobacteria (n = 20), Bacteroidetes (n = 57), Firmicutes (n = 83), Fusobacteria (n = 3), and Proteobacteria (n = 10). Dashed line: metabolite threshold. Box: median, 25th, and 75th percentile; whiskers: Tukey’s method. Bottom panel: The 10 most important features differentiating the five tested phyla. Data are shown as median metabolite log2 fold-change for each phylum; metabolites and phyla are ordered by Ward linkage distance. b, Representative growth curves from two independent experiments, each with n = 3 biological replicates for a subset of Bacteroides spp. using modified Salyer’s Minimal Medium (SMM) with indicated nitrogen source. Legend colors for sole nitrogen source are maintained for panels b-d. c, Representative growth curves of one experiment with n = 5 biological replicates for 60 Bacteroidetes using modified SMM with indicated nitrogen sources. d, Growth curves of wild-type and mutant Bt grown in defined minimal media with either cysteine (top) (one experiment, n = 3 biological replicates) or sodium sulfide (Na2S, bottom) as sole reduced sulfur sources (one experiment, n = 3 biological replicates). e, Amino acid production and consumption levels in gnotobiotic mice mono-associated with Bacteroides thetaiotaomicron (Bt) (one experiment, n = 5 mice). Box: median, 25th, and 75th percentile; whiskers: Tukey’s method). Numeric labels in b and c correspond to the following: 1: B. acidifaciens DSMZ 15896, 2: B. caccae ATCC 43185, 3: B. caccae BEI HM-728, 4: B. cellulosilyticus BEI HM-726, 5: B. cellulosilyticus DSMZ 14838, 6: B. coprophilus DSMZ 18228, 7: B. dorei BEI HM-29, 8: B. dorei BEI HM-717, 9: B. dorei BEI HM-718, 10: B. dorei BEI HM-719, 11: B. dorei DSMZ 17855, 12: B. eggerthii ATCC 27754, 13: B. eggerthii DSMZ 20697, 14: B. finegoldii BEI HM-727, 15: B. finegoldii DSMZ 17565, 16: B. fragilis BEI HM-20, 17: B. fragilis BEI HM-710, 18: B. fragilis BEI HM-711, 19: B. fragilis BEI HM-714, 20: B. fragilis NCTC 9343, 21: B. intestinalis DSMZ 17393, 22: B. ovatus ATCC 8483, 23: B. ovatus BEI HM-222, 24: B. pectinophilus ATCC 43243, 25: B. plebeius DSMZ 17135, 26: B. salyersiae BEI HM-725, 27: B. sp. BEI HM-18, 28: B. sp. BEI HM-189, 29: B. sp. BEI HM-19, 30: B. sp. BEI HM-22, 31: B. sp. BEI HM-23, 32: B. sp. BEI HM-258, 33: B. sp. BEI HM-27, 34: B. sp. BEI HM-28, 35: B. sp. BEI HM-58, 36: B. stercoris ATCC 43183, 37: B. stercoris BEI HM-1036, 38: B. thetaiotaomicron 3730, 39: B. thetaiotaomicron 3731, 40: B. thetaiotaomicron 633, 41: B. thetaiotaomicron 7330, 42: B. thetaiotaomicron 7853, 43: B. thetaiotaomicron 8702, 44: B. thetaiotaomicron 8713, 45: B. thetaiotaomicron 8736, 46: B. thetaiotaomicron 940, 47: B. thetaiotaomicron VPI 5482, 48: B. thetaiotaomicron wh302, 49: B. thetaiotaomicron wh305, 50: B. uniformis ATCC 8492, 51: B. vulgatus ATCC 8482, 52: B. vulgatus BEI HM-720, 53: B. xylanisolvens DSMZ 18836, 54: P. distasonis ATCC 8503, 55: P. distasonis BEI HM-169, 56: P. johnsonii BEI HM-731, 57: P. johnsonii DSMZ 18315, 58: P. merdae ATCC 43184, 59: P. merdae BEI HM-729, 60: P. merdae BEI HM-730.
