Fig. 3, Discovery of nitrogen assimilation strategies in Bacteroides and novel gene-phenotype relationships.
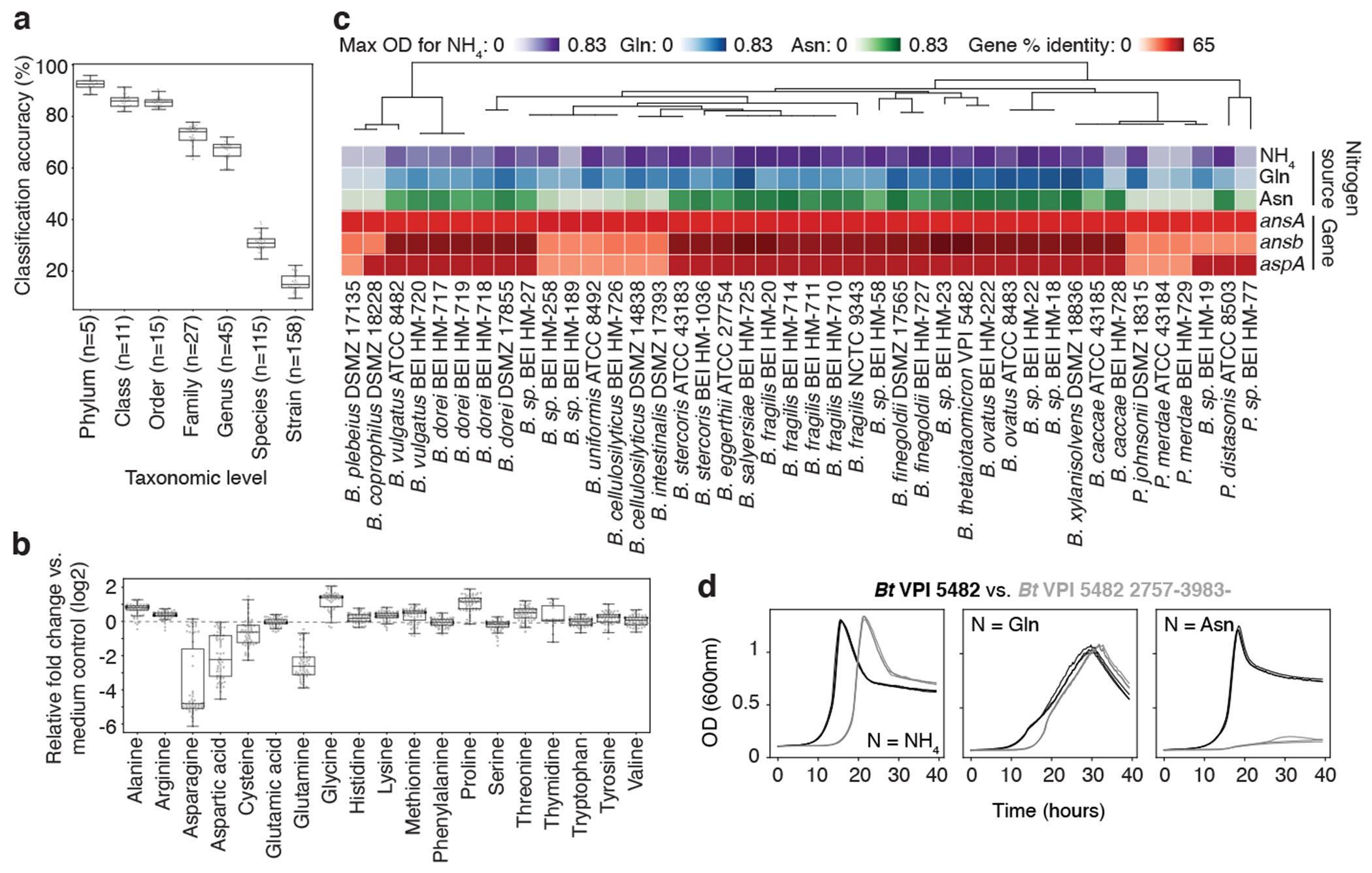
a, Classification accuracy of Random Forest (RF) models at each taxonomic level, based on metabolomic profiles of 158 mega-medium grown bacterial strains from one to three independent experiments, each with n ≥ 3 biological replicates. Phylum (n = 5), Class (n = 11), Order (n = 15), Family (n = 27), Genus (n = 45), Species (n = 115), and Strain (n = 158). b, Amino acid production or consumption levels by Bacteroidetes strains from one to three independent experiments, each with n ≥ 3 biological replicates. Only uniquely detected (non-coeluting) amino acids are shown. a, b, Boxes: median, 25th, and 75th percentile; whiskers: Tukey’s method. c, Phylogenetic tree of Bacteroidetes strains, growth curve max optical density (OD600nm), and percentage of protein sequence identity for E. coli asparagine-consuming, ammonium-liberating enzymes. Nitrogen sources: ammonia (NH4), Glutamine (Gln), Asparagine (Asn). d, Representative growth curves of wild-type and mutant Bt (2757-3983-) in modified Salyer’s Minimal Medium from one experiment with n = 3 biological replicates. Nitrogen sources as in c: NH4, Gln, Asn.
