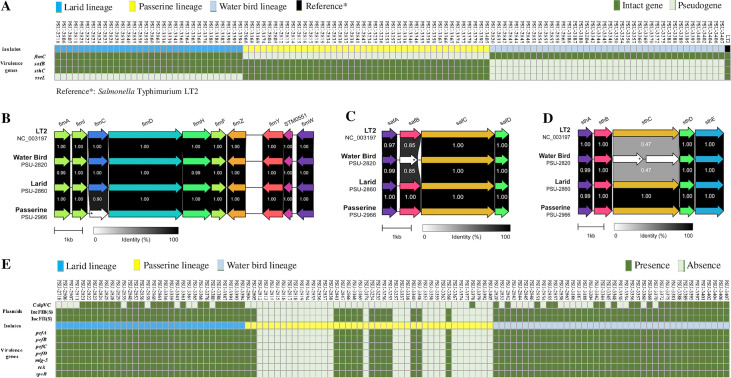
FIG 4.
Chromosomal and plasmid-associated virulence gene signatures that differ among the larid, passerine, and water bird lineages. (A) Distribution of chromosomal virulence gene signatures (fimC, safB, sthC, and sseL) among the larid, passerine, and water bird lineages. (B–D) Organization of the fimbrial operons containing fimC, safB, and sthC in isolates from the larid, passerine, and water bird lineages, and reference strain S. Typhimurium LT2. (E) Distribution of plasmids and plasmid virulence genes among the larid, passerine, and water bird lineages. In the heatmaps (A) and (E): light green = pseudogene in (A), and absence of the gene or plasmid in (E); dark green = intact gene in (A), and presence of the gene or plasmid in (E); blue = isolates from larid lineage; yellow = isolates from passerine lineage; light blue = isolates from water bird lineage; dark = reference strain (S. Typhimurium LT2). Isolate names are represented on the x axis, virulence genes and plasmid names are represented on the y axis. Complete data used to generate the heatmaps is given in Data Set S3. In the fimbrial operons (B), (C), and (D), the pseudogenes are marked as white arrows, and the locations of indels in the pseudogenes are represented by asterisks.