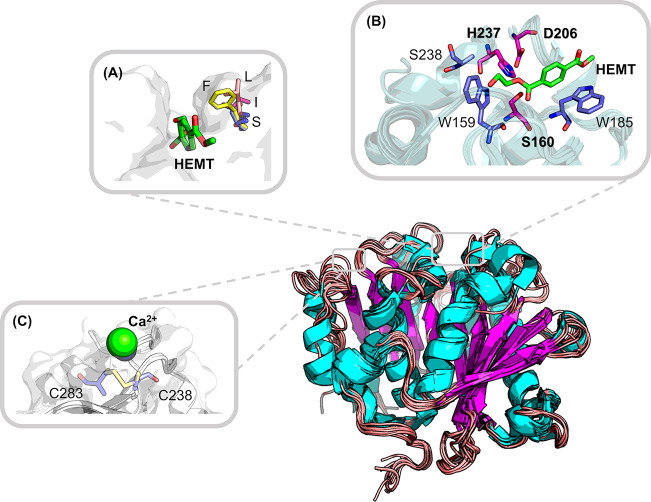
Figure 2.
Frequently reported mutation hot spots illustrated on the superposed crystal structures of known bacterial PET hydrolases. Backbones shown in the cartoon are derived from the structural superposition with selected homologous enzymes. The catalytic triad (S160, D206, and H237) as well as two aromatic residues (W159 and W185) are involved in the interaction with the monomer analogue 1-(2-hydroxyethyl)-4-methyl terephthalate (HEMT) (A, B) based on the IsPETase structure (PDB ID: 5XH3; the numbering of residues is modified consistently with other structures solved later for easy comprehension). (A) One frequently reported mutation hotspot equivalent to S209 (B) in IsPETase can adopt various residues which might influence the widths of the binding pocket:65,90 F found in 4CG1, 4EB0, and 7OSB and also conserved in many other PET hydrolases; S found in the IsPETase structure 5XH3; I found in an LCC mutant 6THT; L found in another PET hydrolase 7CUV. (C) One of the putative Ca2+ binding sites revealed by cocrystallized structures such as 4WFJ, 5LUL, and 5ZNO can be replaced by a disulfide bridge (6THT, 7CTS, and 7CTR) to thermostabilize several PET hydrolases.