FIG 4.
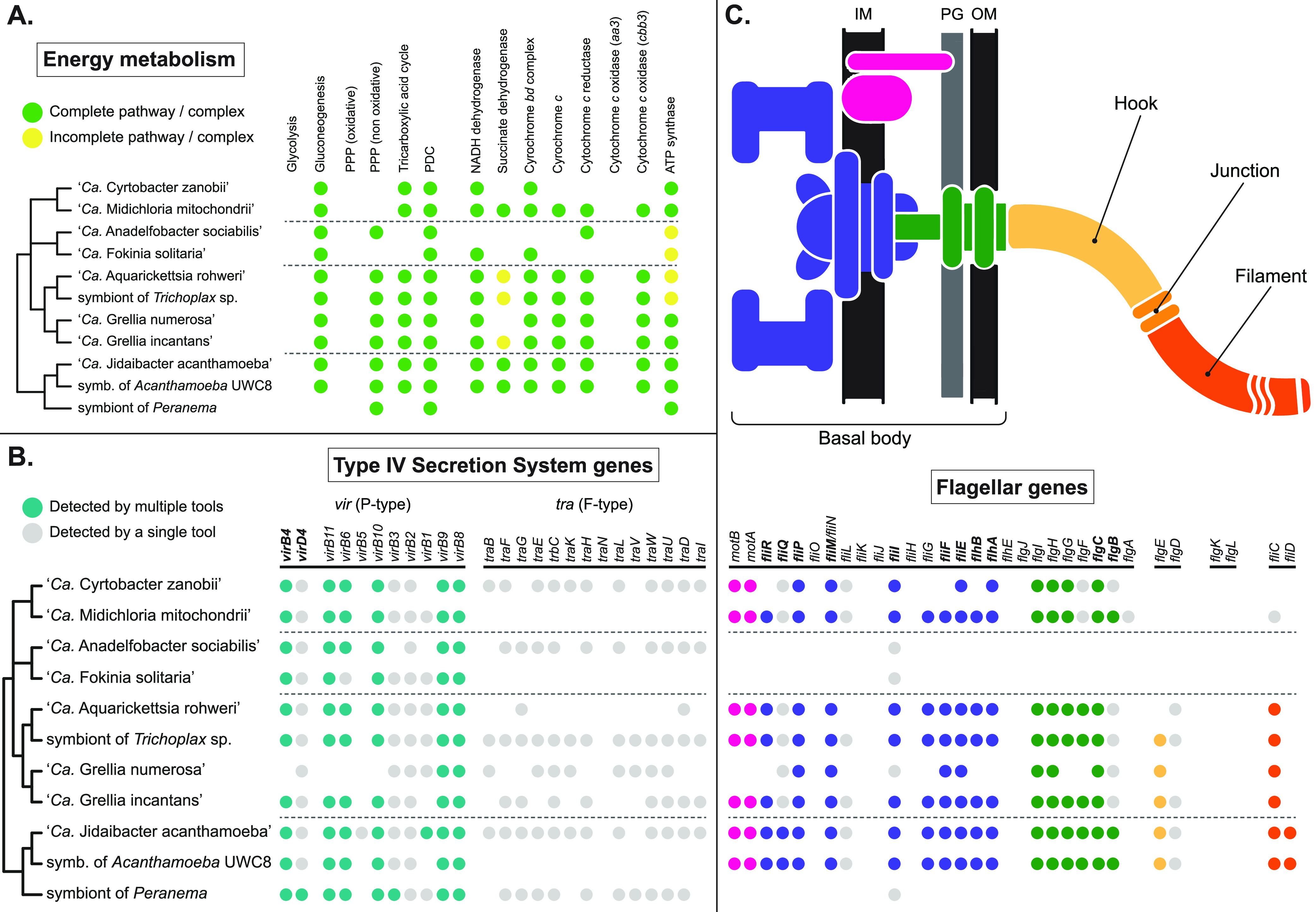
Selected functional traits mapped on the consensus phylogeny of “Ca. Midichloriaceae.” Dashed lines separate the four clades identified in Fig. 2. (A) Energy metabolism-related genes, complexes, and pathways, as annotated by the KEGG Automatic Annotation System (KAAS) and, for oxidative phosphorylation, Prokka. Pathways and complexes were considered complete (green circles) when all essential genes known to be exclusively involved in that pathway/complex were detected, possibly incomplete (yellow circles) when at least half but not all such genes were detected, and likely absent (no circle) otherwise. PPP, pentose phosphate pathway; PDC, pyruvate dehydrogenase complex. (B) Genes involved in T4SS, as annotated by KAAS, Prokka, and TXSScan. Teal circles mark genes detected by at least two analytical tools; gray circles mark genes detected by only one of the three (usually TXSScan, which is optimized for the detection of T4SS genes; see also Fig. S1). Names of genes considered mandatory (73) are in bold. (C) Flagellum and flagellar assembly-related genes, annotated and displayed as in panel B, but with different colors used for different parts of the flagellum, as shown in the schematic drawing above the table. IM, internal membrane; PG, peptidoglycan layer; OM, outer membrane. More details on the functional analysis are shown in Table S2.
