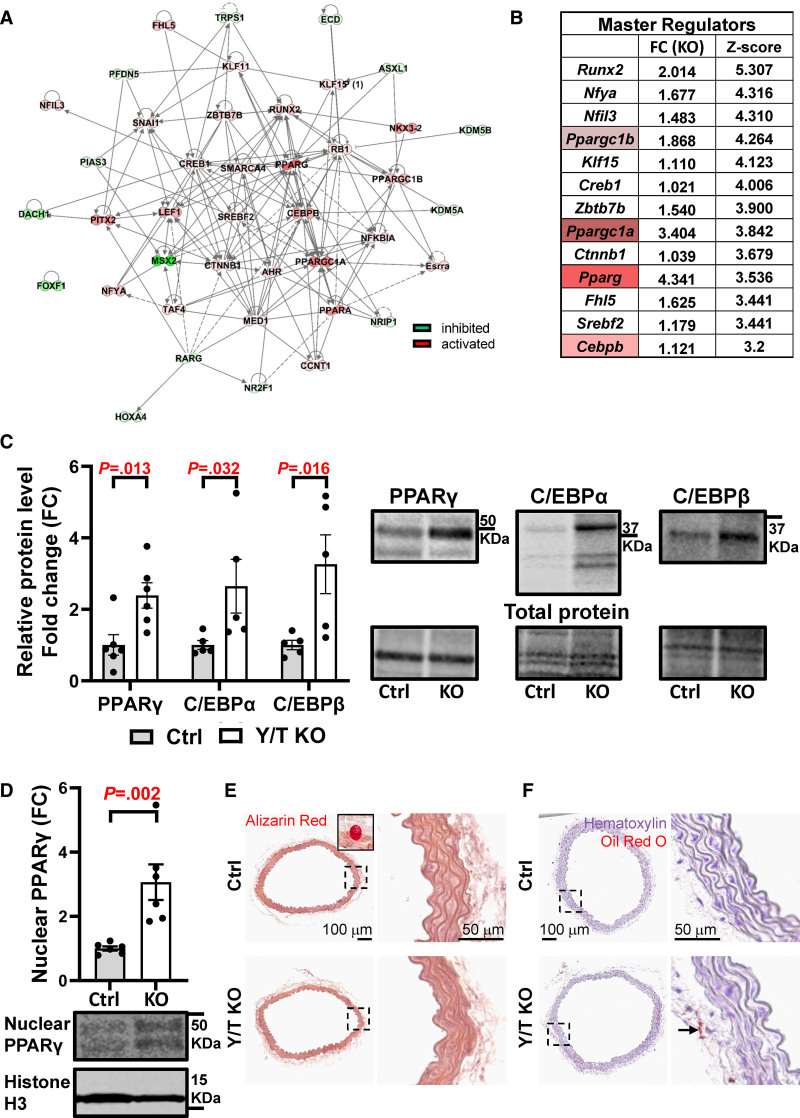
Figure 5.
Upregulation of transcription factors that are involved in adipogenic differentiation: PPARγ (peroxisome proliferator activated receptor gamma), C/EBPα (CCAAT enhancer binding protein alpha), and C/EBPβ (CCAAT enhancer binding protein beta). A, Network of the predicted master regulators that are involved in transcription regulation. The interactions between the nodes are based on Qiagen ingenuity knowledge base, where the solid and dashed lines represent direct and indirect interaction, respectively. The intensity of coloring is based on the magnitude of Z score values ranging from strong inhibition (dark green) to strong activation (dark red). B, The top 13 master regulators and their fold change and Z score, which reflects their activation status. C, Quantification of Western blot analysis of PPARγ, C/EPBα, and C/EPBβ and representative blots to the right (Ctrl [control], n=5–6; Y/T KO [YAP/TAZ (yes-associated protein 1/WW domain containing transcription regulator 1) knockout], n=5–6). D, Quantification and representative blot showing PPARγ in the nuclear fraction and histone H3 as nuclear marker (Ctrl, n=6; Y/T KO, n=6). E, Alizarin Red staining for calcium deposits detection in aortae of Ctrl and Y/T KO mice. Inset represents positive Ctrl (bone). F, Assessment of lipid accumulation in the aorta by Oil Red O staining, positive staining in perivascular fat (arrow). Detailed legend description for IPA network is available online: https://qiagen.secure.force.com/KnowledgeBase/articles/Knowledge/Legend. All data are presented as mean±SEM. Ctrl indicates control; DAPI, 4′,6-diamidino-2-phenylindole; FC, fold change; and KO, knockout.