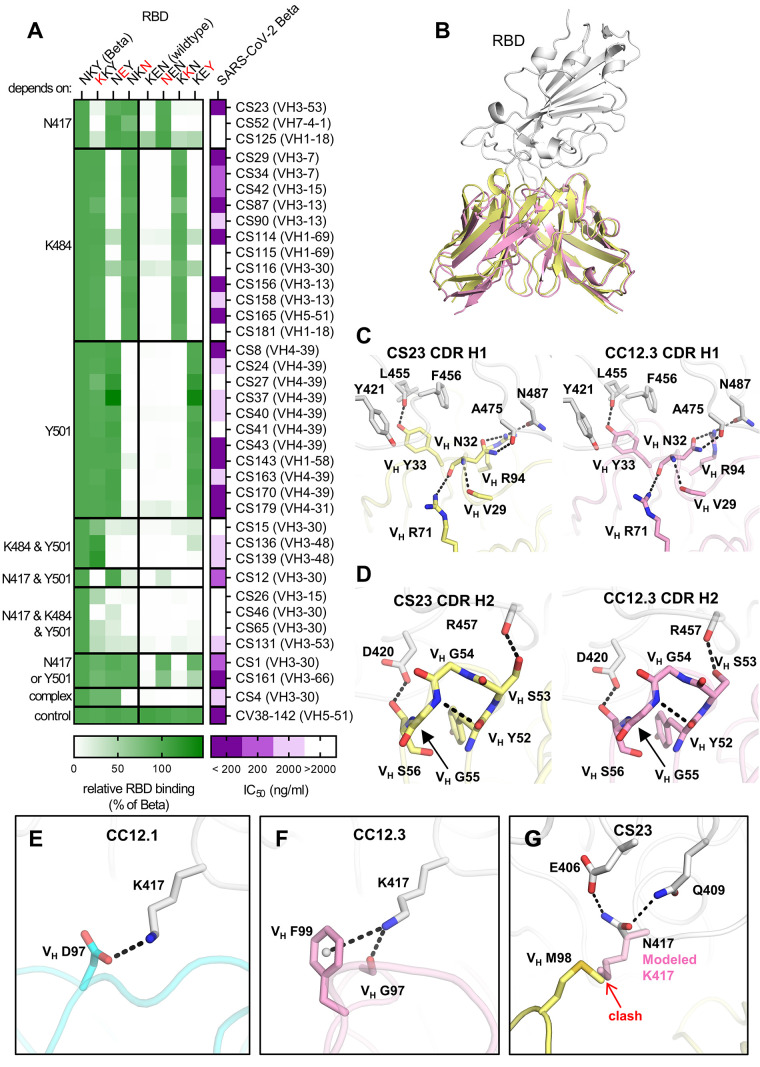
Fig. 3. Binding, neutralization and structures of Beta-specific antibodies.
(A) Neutralization of indicated Beta-specific mAbs against authentic Beta virus is shown in purple. Binding to single point mutant RBD constructs with the indicated amino-acid residues at positions 417, 484 and 501 is shown in green, normalized to RBD Beta. (B to G) Structural comparison of VH3-53 mAbs between Beta-specific CS23 and wildtype-specific CC12.1 and CC12.3. (B) CC12.3 and CS23 adopt the same binding mode. The crystal structure of CC12.3 (pink) in complex with RBD (wildtype) was superimposed onto CS23 (yellow) in complex with RBD (Beta). Only the variable domains of the antibodies are shown for clarity. A small local conformational difference was observed between CS23-bound RBD Beta and CC12.3-bound wildtype RBD (191 Cα, RMSD = 0.8 Å). (C and D) Comparison of the (C) CDR H1 (‘NY’ motif) and (D) CDR H2 (‘SGGS’ motif) between CS23 and CC12.3. (E to G) Structures of CDR H3 of (E) CC12.1, (F) CC12.3, and (G) CS23. A modeled side chain of K417 is shown as transparent pink sticks, which would be unfavorable for binding to CS23, where VH M98 occupies this pocket. Structures of CC12.1 (PDB 6XC3, cyan), CC12.3 (PDB 6XC4, pink), and CS23 (this study, yellow) are used throughout this figure, and the RBD is shown in white. Hydrogen bonds, salt bridges or cation-π bonds are represented by black dashed lines.