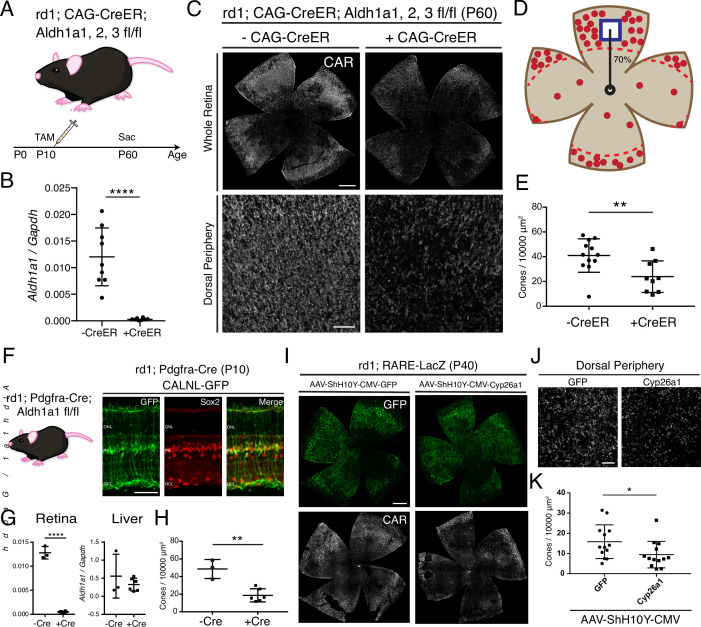
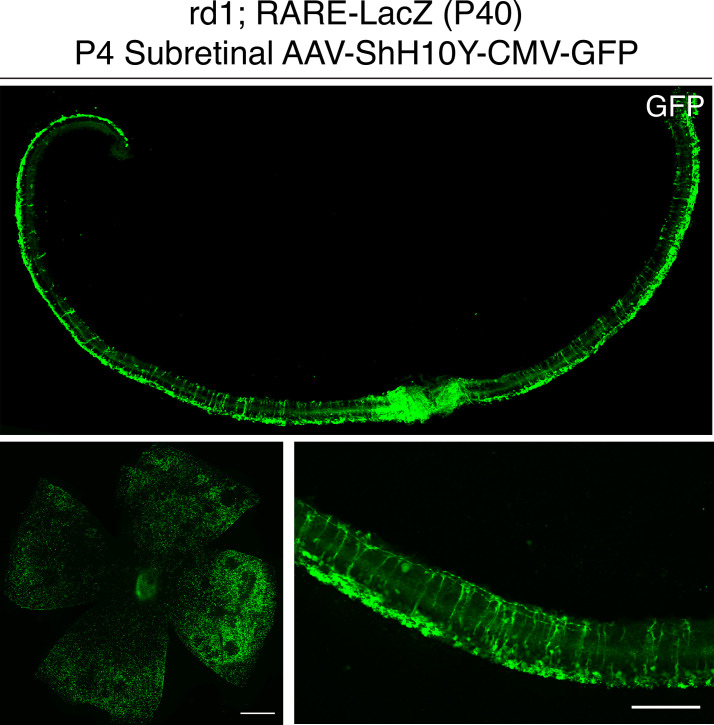
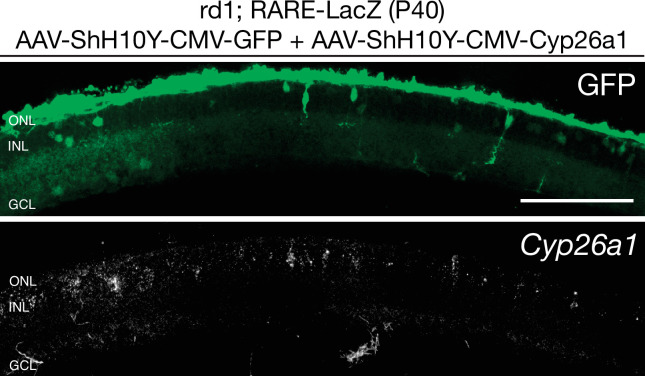
Figure 2. RA is necessary for peripheral cone survival.
(A) Schematic of the experimental design. Aldh Flox mice were injected with tamoxifen to create a conditional knockout (cKO) of the RA synthetic enzymes at P10, and the retinas were harvested at P60. (B) Relative expression level of Aldh1a1 in retinas with or without CAG-CreER (-Cre: n = 9,+ Cre: n = 8, Student’s two-tailed T test, p < 0.0001). (C) IHC against CAR in P60 Aldh Flox flatmounts showing the entire retina (top row) and the dorsal periphery (bottom row) with or without CAG-CreER. (D) Schematic of the area of cone quantification in the dorsal retina. Red dots represent surviving cones, and the blue box is the area of quantification. (E) Quantification of CAR+ cones in the sampled area of Aldh Flox retinas with or without CAG-CreER (-Cre: n = 12,+ Cre: n = 9, Student’s two-tailed T test, p = 0.0086). (F) Validation of MG-specific Aldh1a1 cKO mouse line. IHC against GFP and Sox2 in P10 rd1; Pdgfra-Cre retinas electroporated with a Cre-dependent plasmid, CALNL-GFP. (G) Relative expression level of Aldh1a1 in retinas and liver with or without Pdgfra-Cre (Retina: -Cre: n = 3,+ Cre: n = 6, Student’s two-tailed T test, p < 0.0001; Liver: -Cre: n = 3,+ Cre: n = 6, Student’s two-tailed T test, p = 0.3893). (H) Quantification of CAR+ cones in the sampled area of Aldh Flox retinas with or without Pdgfra-Cre (-Cre: n = 3,+ Cre: n = 6, Student’s two-tailed T test, p = 0.0016). (I) IHC against GFP (top row) and CAR (bottom row) in P40 rd1; RARE-LacZ flatmounts resulting from infection with AAV-ShH10Y-CMV-GFP (left column) or AAV-ShH10Y-CMV-Cyp26a1+ AAV-ShH10Y-CMV-GFP (right column). (J) Insets of the dorsal peripheral regions in both groups. (K) Quantification of CAR+ cones in the sampled area of infected retinas (GFP: n = 13, Cyp26a1: n = 13, Mann-Whitney test, p = 0.0441). ONL, Outer Nuclear Layer; INL, Inner Nuclear Layer; GCL, Ganglion Cell Layer. Scale bars; 500 µm (C top panels, I), 50 µm (F), 100 µm (C bottom panels, J). All results are expressed as the Mean ± SD. *p < 0.05, **p < 0.01, ****p < 0.0001.