Figure 7.
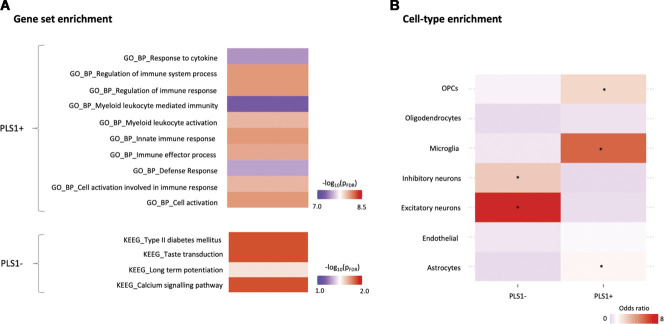
Gene set and cell-type enrichment analyses of the top genes associated with cross-condition changes in regional morphometric similarity during chronic pain. In panel A, we present the results of gene set enrichment analyses on the top genes positively (PLS1+) and negatively (PLS1−) associated with cross-condition changes in regional morphometric similarity during chronic pain, as implemented in the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) platform. For PLS1+, we present the top 10 Gene Ontology (GO)—biological pathways terms for which we found significant enrichment, ranked by P-value after FDR correction. For PLS1−, none of the GO terms survived correction, but we found significant enrichment for 4 terms from the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways. Colour scale indicates –log(PFDR). The full results of the FUMA analysis can be found in supplementary data (available at http://links.lww.com/PAIN/B509, http://links.lww.com/PAIN/B510, http://links.lww.com/PAIN/B511). In panel B, we present the results of a cell-type enrichment analysis, where we investigated whether PLS1+ and PLS1− include overrepresentation of genes typically expressed in specific brain cell types. Enrichment was quantified using odds ratio (OR) and significance calculated with a Fisher exact test. Colour scale indicates OR. Higher ORs (in red) indicate higher enrichment in genes of a certain cell class. The asterisk denotes cell classes for which we found significant enrichment, after correcting for the number of cell types tested (*PFDR < 0.05). FDR, false discovery rate; OPCs, oligodendrocyte precursor cells.